1ELS
 
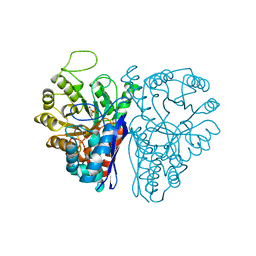 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
1EE6
 
 | | CRYSTAL STRUCTURE OF PECTATE LYASE FROM BACILLUS SP. STRAIN KSM-P15. | | Descriptor: | CALCIUM ION, PECTATE LYASE | | Authors: | Akita, M, Suzuki, A, Kobayashi, T, Ito, S, Yamane, T. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first structure of pectate lyase belonging to polysaccharide lyase family 3.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6JEB
 
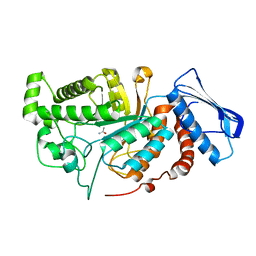 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
1EEQ
 
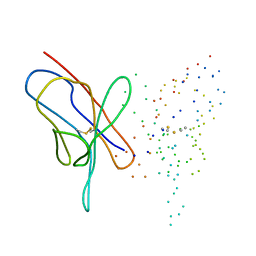 | | M4L/Y(27D)D/T94H Mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Raffen, R, Dieckman, L, Boogaard, C, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increasing protein stability by polar surface residues: domain-wide consequences of interactions within a loop.
Biophys.J., 82, 2002
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
1EFH
 
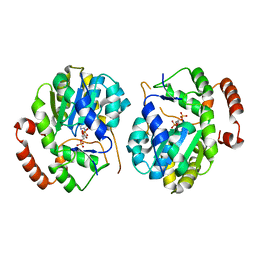 | |
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | Descriptor: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-12-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.61 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
1EG4
 
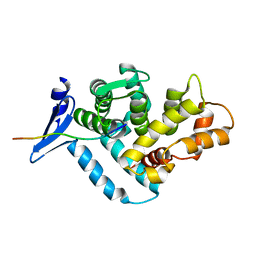 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | BETA-DYSTROGLYCAN, DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
1EH4
 
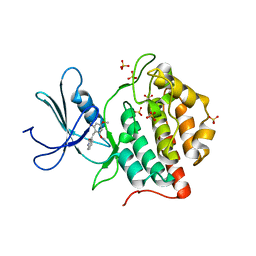 | | BINARY COMPLEX OF CASEIN KINASE-1 FROM S. POMBE WITH AN ATP COMPETITIVE INHIBITOR, IC261 | | Descriptor: | 3-[(2,4,6-TRIMETHOXY-PHENYL)-METHYLENE]-INDOLIN-2-ONE, CASEIN KINASE-1, SULFATE ION | | Authors: | Mashhoon, N, Demaggio, A.J, Tereshko, V, Bergmeier, S.C, Egli, M, Hoekstra, M.F, Kuret, J. | | Deposit date: | 2000-02-18 | | Release date: | 2001-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Conformation-Selective Casein Kinase-1 Inhibitor
J.Biol.Chem., 275, 2000
|
|
6IEN
 
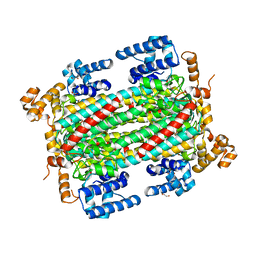 | | Substrate/product bound Argininosuccinate lyase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ARGININOSUCCINATE, ... | | Authors: | Paul, A, Mishra, A, Surolia, A, Vijayan, M. | | Deposit date: | 2018-09-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on M. tuberculosis argininosuccinate lyase and its liganded complex: Insights into catalytic mechanism.
IUBMB Life, 71, 2019
|
|
1EQJ
 
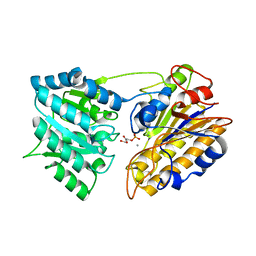 | | CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J.Biol.Chem., 275, 2000
|
|
1EQU
 
 | | TYPE 1 17-BETA HYDROXYSTEROID DEHYDROGENASE EQUILIN COMPLEXED WITH NADP+ | | Descriptor: | EQUILIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (ESTRADIOL 17 BETA-DEHYDROGENASE 1) | | Authors: | Sawicki, M.W, Erman, M, Puranen, T, Vihko, P, Ghosh, D. | | Deposit date: | 1998-12-02 | | Release date: | 1999-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the ternary complex of human 17beta-hydroxysteroid dehydrogenase type 1 with 3-hydroxyestra-1,3,5,7-tetraen-17-one (equilin) and NADP+.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1EMW
 
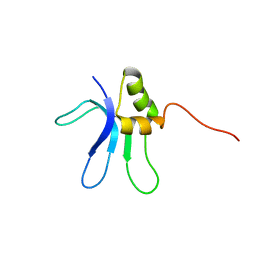 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S16 FROM THERMUS THERMOPHILUS | | Descriptor: | S16 RIBOSOMAL PROTEIN | | Authors: | Allard, P, Rak, A.V, Wimberly, B.T, Clemons Jr, W.M, Kalinin, A, Helgstrand, M, Garber, M.B, Ramakrishnan, V, Hard, T. | | Deposit date: | 2000-03-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Another piece of the ribosome: solution structure of S16 and its location in the 30S subunit.
Structure Fold.Des., 8, 2000
|
|
7UR3
 
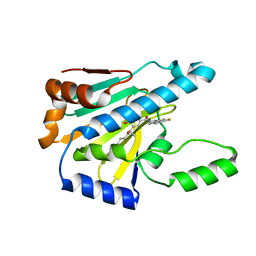 | | Hsp90 alpha inhibitor | | Descriptor: | (5-fluoro-1,3-dihydro-2H-isoindol-2-yl){4-hydroxy-3-[(2S)-2-hydroxy-5-phenylpentan-2-yl]phenyl}methanone, Heat shock protein HSP 90-alpha | | Authors: | Deng, J, Peng, S, Balch, M, Matts, R. | | Deposit date: | 2022-04-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Activity Relationship Study of Tertiary Alcohol Hsp90 alpha-Selective Inhibitors with Novel Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
1EXA
 
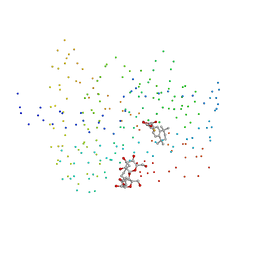 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8UFD
 
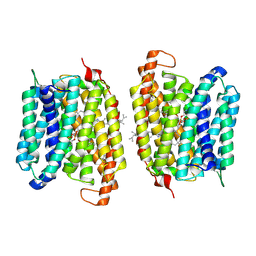 | | Multidrug efflux pump MtEfpA bound with inhibitor BRD8000.3 | | Descriptor: | (1S,3S)-N-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3-(2-methylpropyl)cyclopropane-1-carboxamide, Integral membrane efflux protein EFPA, PHOSPHATIDYLETHANOLAMINE | | Authors: | Wang, S, Liao, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of the Mycobacterium tuberculosis efflux pump EfpA reveal the mechanisms of transport and inhibition.
Nat Commun, 15, 2024
|
|
1EXV
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
7U9I
 
 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
1EKY
 
 | |
1EPR
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EQ7
 
 | |
