2XCN
 
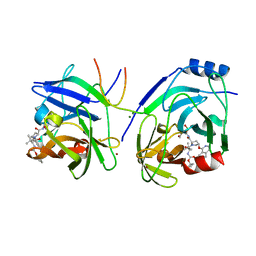 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | MAGNESIUM ION, N-[(CYCLOPENTYLOXY)CARBONYL]-3-METHYL-L-VALYL-(4R)-N-{(1R)-3-HYDROXY-1-[HYDROXY(OXIDO)BORANYL]PROPYL}-4-(ISOQUINOLIN-1-YLOXY)-L-PROLINAMIDE, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4WR5
 
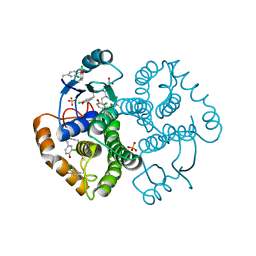 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
8C19
 
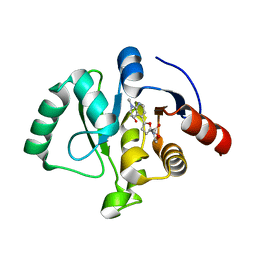 | | SARS-CoV-2 NSP3 macrodomain in complex with 1-methyl-4-[5-(morpholin-4-ylcarbonyl)-2-furyl]-1H-pyrrolo[2,3-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | | Authors: | Schuller, M, Ahel, I. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Development Strategies for SARS-CoV-2 NSP3 Macrodomain Inhibitors.
Pathogens, 12, 2023
|
|
3NN2
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with cyanide | | Descriptor: | CYANIDE ION, Chlorite dismutase, GLYCEROL, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
2XCF
 
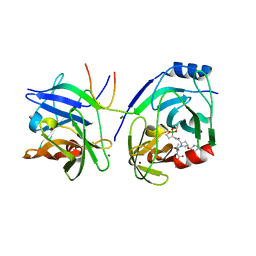 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | CYCLOPENTYL N-[(2S)-1-[(2S,4R)-2-[[(4R)-8-HYDROXY-1,6,10-TRIOXA-5$L^{4}-BORASPIRO[4.5]DECAN-4-YL]CARBAMOYL]-4-ISOQUINOLIN-1-YLOXY-PYRROLIDIN-1-YL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5B7E
 
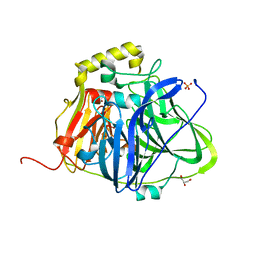 | | Structure of perdeuterated CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5BNW
 
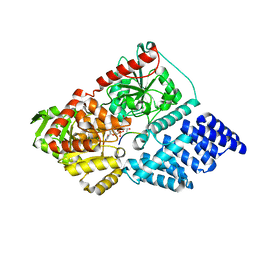 | | The active site of O-GlcNAc transferase imposes constraints on substrate sequence | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, laminB1 residues 179-191 | | Authors: | Pathak, S, Alonso, J, Schimpl, M, Rafie, K, Blair, D.E, Borodkin, V.S, Albarbarawi, O, van Aalten, D.M.F. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1ZK5
 
 | | Escherichia coli F17fG lectin domain complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17G adhesin subunit | | Authors: | Buts, L, Wellens, A, Van Molle, I, De Genst, E, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-02 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of natural variation in bacterial F17G adhesins on crystallization behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZKZ
 
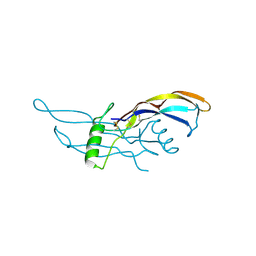 | | Crystal Structure of BMP9 | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Brown, M.A, Zhao, Q, Baker, K.A, Naik, C, Chen, C, Pukac, L, Singh, M, Tsareva, T, Parice, Y, Mahoney, A, Roschke, V, Sanyal, I, Choe, S. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of BMP-9 and functional interactions with pro-region and receptors
J.Biol.Chem., 280, 2005
|
|
2XCS
 
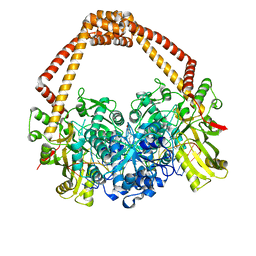 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
8C03
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy12 in outward-facing conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, Solute carrier family 40 member 1 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
4X29
 
 | |
1ZM8
 
 | | Apo Crystal structure of Nuclease A from Anabaena sp. | | Descriptor: | MANGANESE (II) ION, Nuclease, SODIUM ION, ... | | Authors: | Ghosh, M, Meiss, G, Pingoud, A, London, R.E, Pedersen, L.C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Mechanism of Nuclease A, a beta beta alpha Metal Nuclease from Anabaena.
J.Biol.Chem., 280, 2005
|
|
4JD7
 
 | | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate | | Descriptor: | Proline racemase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-24 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group P212121, bound sulfate
To be Published
|
|
2XH1
 
 | | Crystal structure of human KAT II-inhibitor complex | | Descriptor: | (3S)-10-(4-AMINOPIPERAZIN-1-YL)-9-FLUORO-7-HYDROXY-3-METHYL-2,3-DIHYDRO-8H-[1,4]OXAZINO[2,3,4-IJ]QUINOLINE-6-CARBOXYLATE, GLYCEROL, IODIDE ION, ... | | Authors: | Rossi, F, Casazza, V, Garavaglia, S, Sathyasaikumar, K.V, Schwarcz, R, Kojima, S.I, Okuwaki, K, Ono, S.I, Kajii, Y, Rizzi, M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure-Based Selective Targeting of the Pyridoxal 5'-Phsosphate Dependent Enzyme Kynurenine Aminotransferase II for Cognitive Enhancement
J.Med.Chem., 53, 2010
|
|
8BZY
 
 | | Structure of SLC40/ferroportin in complex with vamifeport and synthetic nanobody Sy3 in occluded conformation | | Descriptor: | 2-[2-[2-(1~{H}-benzimidazol-2-yl)ethylamino]ethyl]-~{N}-[(3-fluoranylpyridin-2-yl)methyl]-1,3-oxazole-4-carboxamide, DIUNDECYL PHOSPHATIDYL CHOLINE, Solute carrier family 40 member 1, ... | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
4JDX
 
 | |
4JED
 
 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
8C02
 
 | | Structure of SLC40/ferroportin in complex with synthetic nanobody Sy3 in occluded conformation | | Descriptor: | Solute carrier family 40 member 1, Sybody3 | | Authors: | Lehmann, E.F, Liziczai, M, Drozdzyk, K, Dutzler, R, Manatschal, C. | | Deposit date: | 2022-12-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structures of ferroportin in complex with its specific inhibitor vamifeport.
Elife, 12, 2023
|
|
2WR6
 
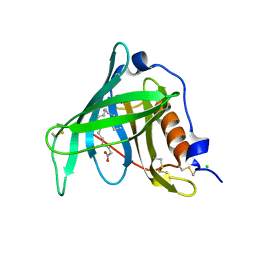 | | Structure of the complex of RBP4 with linoleic acid | | Descriptor: | (11E,13E,15Z)-OCTADECA-11,13,15-TRIENOIC ACID, CHLORIDE ION, RETINOL-BINDING PROTEIN 4 | | Authors: | Huang, H.-J, Nanao, M, Stout, T, Rosen, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Non-Retinoid Compound and Fatty Acids as Ligands for Retinol Binding Protein 4 and Their Implications in Diabetes
To be Published
|
|
4JOI
 
 | | Crystal structure of the human telomeric Stn1-Ten1 complex | | Descriptor: | CST complex subunit STN1, CST complex subunit TEN1 | | Authors: | Bryan, C, Rice, C, Harkisheimer, M, Schultz, D, Skordalakes, E. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the human telomeric stn1-ten1 capping complex.
Plos One, 8, 2013
|
|
5BR5
 
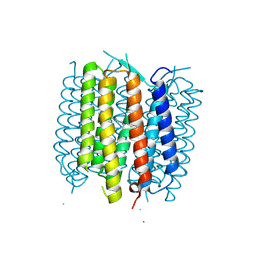 | | Structure of bacteriorhodopsin crystallized from ND-MSP1E3D1 | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nikolaev, M, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-05-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Integral Membrane Proteins Can Be Crystallized Directly from Nanodiscs
Cryst.Growth Des., 17, 2017
|
|
8CEV
 
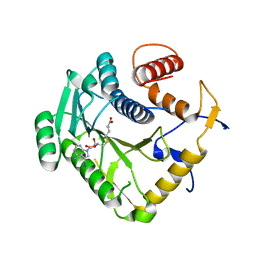 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO1119 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-iodanyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
2WUW
 
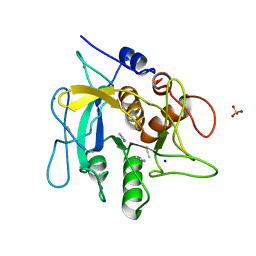 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile (native data) | | Descriptor: | ACETONITRILE, CALCIUM ION, SODIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
2WUV
 
 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile | | Descriptor: | ACETONITRILE, CALCIUM ION, CESIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
