5R13
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 18, DMSO-free | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5FGF
 
 | |
5R1N
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 38, DMSO-free | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6GXA
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 2 | | 分子名称: | (~{E})-3-(2-chlorophenyl)-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | 著者 | Shaik, T.B, Marek, M, Romier, C. | | 登録日 | 2018-06-27 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
5R21
 
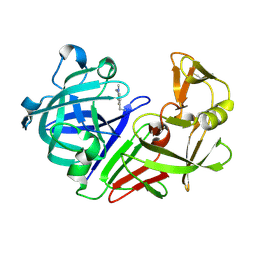 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry E07, DMSO-free | | 分子名称: | 1-(1-methyl-1,2,3,4-tetrahydroquinolin-6-yl)methanamine, Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.047 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8EMB
 
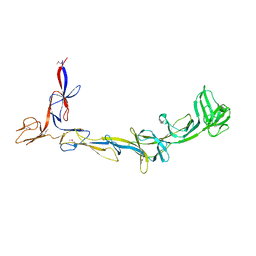 | | X-ray crystal structure of Thermosynechococcus elongatus Si3 domain of RNA polymerase RpoC2 subunit | | 分子名称: | DNA-directed RNA polymerase subunit beta' | | 著者 | Murakami, K.S, Imashimizu, M, Qayyum, M.Z, Vishwakarma, R.K, Yuzenkova, Y. | | 登録日 | 2022-09-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8EP9
 
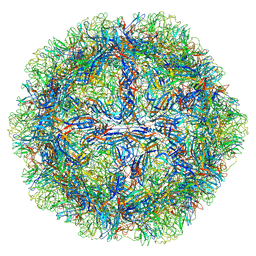 | |
5R2H
 
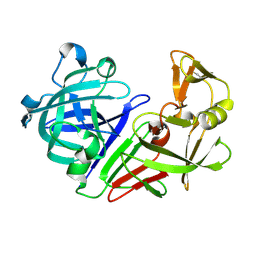 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 04, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.018 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2W
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 20, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.285 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3A
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 34, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.088 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
5R3S
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 52, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.048 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | 分子名称: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | 著者 | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | 登録日 | 2021-03-05 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
6GXU
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 3 | | 分子名称: | (~{E})-3-[2-(4-chlorophenyl)sulfanylphenyl]-~{N}-oxidanyl-prop-2-enamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | 著者 | Shaik, T.B, Marek, M, Romier, C. | | 登録日 | 2018-06-27 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.917 Å) | | 主引用文献 | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
5JD5
 
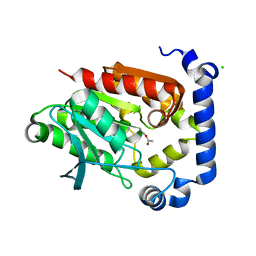 | | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy | | 分子名称: | CHLORIDE ION, MGS-MilE3 | | 著者 | Stogios, P.J, Xu, X, Cui, H, Martinez-Martinez, M, Chernikova, T.N, Golyshin, P.N, Yakimov, M.M, Ferrer, M, Savchenko, A. | | 登録日 | 2016-04-15 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy
To Be Published
|
|
2POE
 
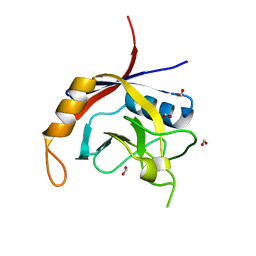 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | 分子名称: | Cyclophilin-like protein, putative, FORMIC ACID | | 著者 | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2007-04-26 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
3OUE
 
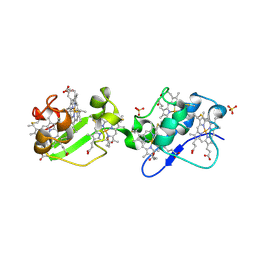 | |
7L0R
 
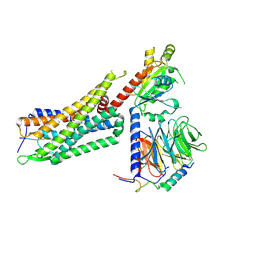 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | 登録日 | 2020-12-12 | | 公開日 | 2021-01-06 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JK8
 
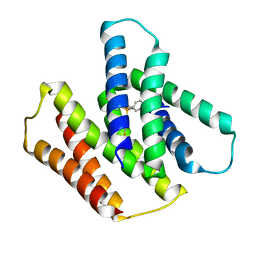 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | 分子名称: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | 著者 | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | 登録日 | 2020-07-27 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
7L0P
 
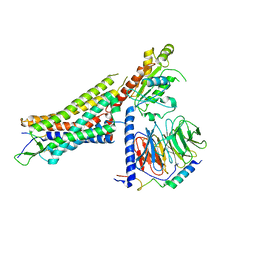 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | 登録日 | 2020-12-12 | | 公開日 | 2021-01-06 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
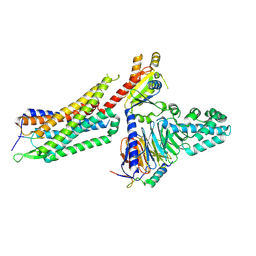 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | 登録日 | 2020-12-12 | | 公開日 | 2021-01-06 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L6B
 
 | |
5V9T
 
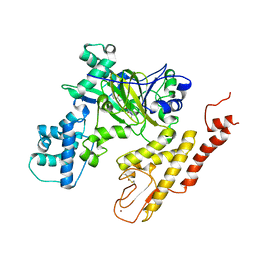 | | Crystal structure of selective pyrrolidine amide KDM5a inhibitor N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide (compound 48) | | 分子名称: | Lysine-specific demethylase 5A, N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide, NICKEL (II) ION, ... | | 著者 | Kiefer, J.R, Liang, J, Vinogradova, M. | | 登録日 | 2017-03-23 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VAS
 
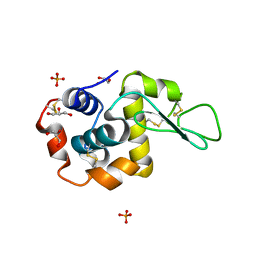 | | Pekin duck egg lysozyme isoform III (DEL-III), orthorhombic form | | 分子名称: | GLYCEROL, Lysozyme, PHOSPHATE ION | | 著者 | Christie, M, Christ, D, Langley, D.B. | | 登録日 | 2017-03-27 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
