2RCY
 
 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-20 | | 公開日 | 2007-10-23 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | 分子名称: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | 著者 | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | 登録日 | 2007-09-24 | | 公開日 | 2008-03-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.1 Å) | | 主引用文献 | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
5UK3
 
 | |
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | 著者 | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | 登録日 | 2002-12-04 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
5UQH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | 分子名称: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-08 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
1IX3
 
 | | Crystal Structure of Rat Heme Oxygenase-1 in complex with Heme bound to Cyanide | | 分子名称: | CYANIDE ION, HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sugishima, M, Sakamoto, H, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | 登録日 | 2002-06-10 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
7B36
 
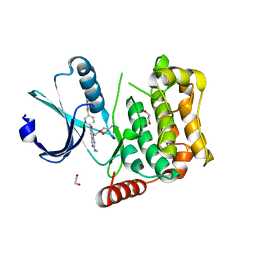 | | MST4 in complex with compound G-5555 | | 分子名称: | 1,2-ETHANEDIOL, 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, ... | | 著者 | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-11-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.10681081 Å) | | 主引用文献 | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | 著者 | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | 登録日 | 2023-03-13 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Ras-like GTPases mutants structure
To be published
|
|
4RG2
 
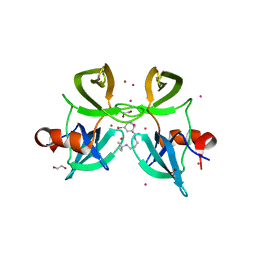 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | 著者 | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-09-29 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | 著者 | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-08-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
1J2N
 
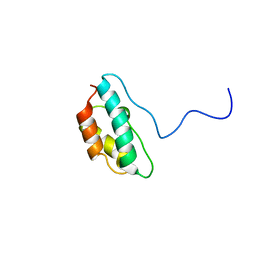 | | Solution structure of CPI-17(22-120) T38D | | 分子名称: | 17-kDa PKC-potentiated inhibitory protein of PP1 | | 著者 | Ohki, S, Eto, M, Shimizu, M, Takada, R, Brautigan, D.L, Kainosho, M. | | 登録日 | 2003-01-07 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Distinctive Solution Conformation of Phosphatase Inhibitor CPI-17 Substituted with Aspartate at the Phosphorylation-site Threonine Residue
J.Mol.Biol., 326, 2003
|
|
6RWJ
 
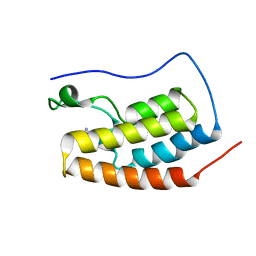 | | Crystal Structure of BRD4(1) bound to inhibitor BUG0 (6) | | 分子名称: | Bromodomain-containing protein 4, ~{N},3-dimethyl-4-oxidanylidene-5,6,7,8-tetrahydro-2~{H}-cyclohepta[c]pyrrole-1-carboxamide | | 著者 | Huegle, M. | | 登録日 | 2019-06-05 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines.
J.Med.Chem., 63, 2020
|
|
6FJG
 
 | |
5GU5
 
 | |
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | 分子名称: | TRAF3-interacting protein 1 | | 著者 | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2023-08-08 | | 公開日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
7AXA
 
 | | Crystal structure of the hPXR-LBD in complex with clotrimazole | | 分子名称: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Delfosse, V, Granell, M, Blanc, P, Bourguet, W. | | 登録日 | 2020-11-09 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VIM
 
 | |
7DQC
 
 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | 分子名称: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | 著者 | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | 登録日 | 2020-12-23 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.706 Å) | | 主引用文献 | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
5K5S
 
 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | 著者 | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | 登録日 | 2016-05-23 | | 公開日 | 2016-08-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
7L0Y
 
 | |
8BR0
 
 | | ExoY Nucleotidyl Cyclase domain from Vibrio nigripulchritudo MARTX toxin (residue Q3455 to L3863) in complex with 3'deoxyCTP and two manganese cations bound to Latrunculin-B-ADP-Mn-actin | | 分子名称: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | 著者 | Texeira-Nuns, M, Retailleau, P, Renault, L. | | 登録日 | 2022-11-22 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.218 Å) | | 主引用文献 | Functional and structural insights into the multi-step activation and catalytic mechanism of bacterial ExoY nucleotidyl cyclase toxins bound to actin-profilin.
Plos Pathog., 19, 2023
|
|
2R9O
 
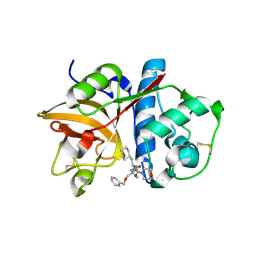 | | Cathepsin S complexed with Compound 8 | | 分子名称: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | 著者 | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | 登録日 | 2007-09-13 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
2R9N
 
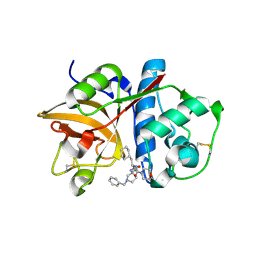 | | Cathepsin S complexed with Compound 26 | | 分子名称: | Cathepsin S, N-[(1S)-2-{[(3S)-1-benzyl-3-cyanopyrrolidin-3-yl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | 著者 | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | 登録日 | 2007-09-13 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
8JZ8
 
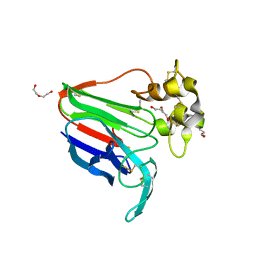 | | Subatomic structure of orthorhombic thaumatin at 0.89 Angstroms | | 分子名称: | DI(HYDROXYETHYL)ETHER, Thaumatin I | | 著者 | Masuda, T, Suzuki, M, Yamasaki, M, Mikami, B. | | 登録日 | 2023-07-04 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (0.89 Å) | | 主引用文献 | Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Biochem.Biophys.Res.Commun., 703, 2024
|
|
7KN5
 
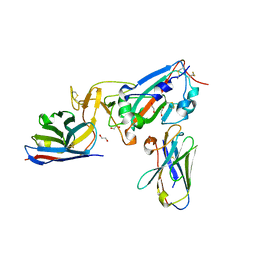 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | 登録日 | 2020-11-04 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
