8SPF
 
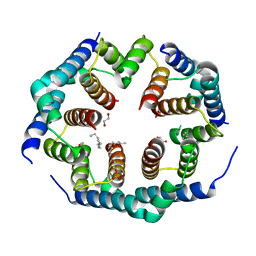 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with 2-stearoyl lysoPC | | 分子名称: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DODECANE, ... | | 著者 | Cowan, A.D, Miller, M.S, Czabotar, P.E, Colman, P.M. | | 登録日 | 2023-05-03 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SRX
 
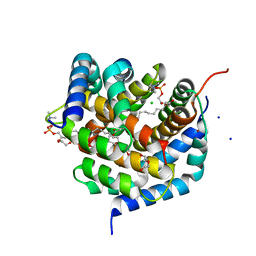 | | Crystal structure of BAK-BAX heterodimer with lysoPC | | 分子名称: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2 homologous antagonist/killer, ... | | 著者 | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | 登録日 | 2023-05-07 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SRY
 
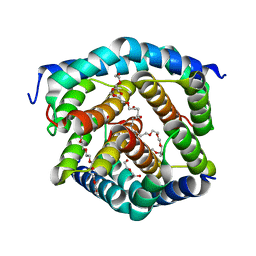 | | Crystal structure of BAK-BAX heterodimer with C12E8 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, ... | | 著者 | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | 登録日 | 2023-05-08 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SPE
 
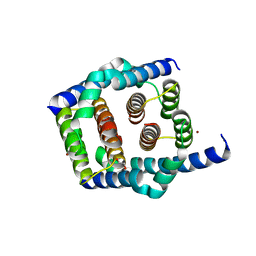 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | 分子名称: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2023-05-03 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
6FTT
 
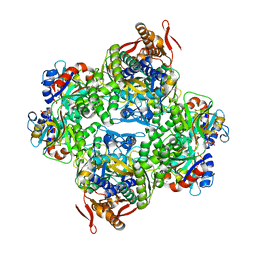 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRPP | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | 著者 | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | 登録日 | 2018-02-23 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FUA
 
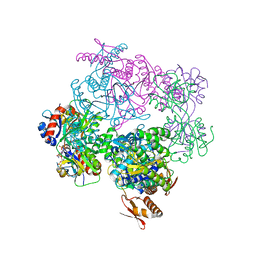 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRPP and ADP | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ATP phosphoribosyltransferase, ... | | 著者 | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | 登録日 | 2018-02-26 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
4OMJ
 
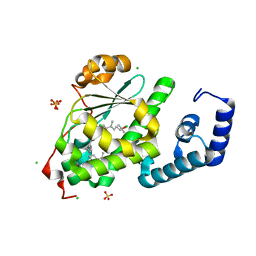 | | Crystal structure of SPF bound to 2,3-oxidosqualene | | 分子名称: | (3S)-2,2-dimethyl-3-[(3E,7E,11E,15E)-3,7,12,16,20-pentamethylhenicosa-3,7,11,15,19-pentaen-1-yl]oxirane, CHLORIDE ION, SEC14-like protein 2, ... | | 著者 | Christen, M, Marcaida, M.J, Lamprakis, C, Cascella, M, Stocker, A. | | 登録日 | 2014-01-27 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights on cholesterol endosynthesis: Binding of squalene and 2,3-oxidosqualene to supernatant protein factor.
J.Struct.Biol., 190, 2015
|
|
6Z96
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) soaked with 4-thiouracil (12.65 keV, 7.125 keV (Fe-SAD) and 6.5 keV (S-SAD) data) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, HYDROSULFURIC ACID, ... | | 著者 | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | 登録日 | 2020-06-03 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.327 Å) | | 主引用文献 | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6FYM
 
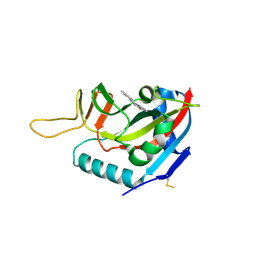 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor ITK1 | | 分子名称: | 7,8-dimethyl-2-(pyrimidin-2-ylsulfanylmethyl)-3~{H}-quinazolin-4-one, NITRATE ION, Poly [ADP-ribose] polymerase 14 | | 著者 | Karlberg, T, Thorsell, A.G, Kirby, I.T, Sreenivasan, R, Cohen, M.S, Schuler, H. | | 登録日 | 2018-03-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity.
Cell Chem Biol, 25, 2018
|
|
107L
 
 | |
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | 著者 | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
108L
 
 | |
109L
 
 | |
112L
 
 | |
110L
 
 | |
111L
 
 | |
113L
 
 | |
3BUG
 
 | |
115L
 
 | |
114L
 
 | |
119L
 
 | |
118L
 
 | |
120L
 
 | |
6G7C
 
 | | Nt2-CTD domains of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | 分子名称: | ImpA-related domain protein | | 著者 | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | 登録日 | 2018-04-05 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
122L
 
 | |
