1HVV
 
 | |
2AZD
 
 | |
2ATV
 
 | | The crystal structure of human RERG in the GDP bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-like estrogen-regulated growth inhibitor | | Authors: | Turnbull, A.P, Salah, E, Schoch, G, Elkins, J, Burgess, N, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human RERG in the GDP bound state
To be Published
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
1LM0
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
2B5U
 
 | | Crystal Structure Of Colicin E3 V206C Mutant In Complex With Its Immunity Protein | | Descriptor: | CITRIC ACID, Colicin E3, Colicin E3 immunity protein | | Authors: | Nallini Vijayarangan, A, Nithianantham, S, Nan, W, Jakes, K, Shoham, M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Of Colicin E3 In Complex With Its Immunity Protein
TO BE PUBLISHED
|
|
2B8A
 
 | | High Resolution Structure of the HDGF PWWP Domain | | Descriptor: | Hepatoma-derived growth factor | | Authors: | Lukasik, S.M, Cierpicki, T, Borloz, M, Grembecka, J, Everett, A, Bushweller, J.H. | | Deposit date: | 2005-10-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the HDGF PWWP domain: a potential DNA binding domain.
Protein Sci., 15, 2006
|
|
2AHQ
 
 | |
2ARP
 
 | | Activin A in complex with Fs12 fragment of follistatin | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Follistatin, GLYCEROL, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-21 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
2AS0
 
 | | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | | Descriptor: | hypothetical protein PH1915 | | Authors: | Sun, W, Xu, X, Pavlova, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a novel SAM-dependent methyltransferase PH1915 from Pyrococcus horikoshii.
Protein Sci., 14, 2005
|
|
2B5I
 
 | | cytokine receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common gamma chain, ... | | Authors: | Wang, X, Rickert, M, Garcia, K.C. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors.
Science, 310, 2005
|
|
2AUN
 
 | |
6WGU
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase | | Descriptor: | Corrinoid adenosyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2AZS
 
 | | NMR structure of the N-terminal SH3 domain of Drk (calculated without NOE restraints) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
1MO9
 
 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE COMPLEXED WITH 2-KETOPROPYL COENZYME M | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-08 | | Release date: | 2002-11-27 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
2AMC
 
 | | Crystal structure of Phenylalanyl-tRNA synthetase complexed with L-tyrosine | | Descriptor: | MAGNESIUM ION, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Kotik-Kogan, O, Moor, N, Tworowski, D, Safro, M. | | Deposit date: | 2005-08-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Discrimination of L-Phenylalanine from L-Tyrosine by Phenylalanyl-tRNA Synthetase
Structure, 13, 2005
|
|
2AQZ
 
 | | Crystal structure of FGF-1, S17T/N18T/G19 deletion mutant | | Descriptor: | Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
1T2V
 
 | | Structural basis of phospho-peptide recognition by the BRCT domain of BRCA1, structure with phosphopeptide | | Descriptor: | BRCTide-7PS, Breast cancer type 1 susceptibility protein | | Authors: | Williams, R.S, Lee, M.S, Hau, D.D, Glover, J.N.M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Nat.Struct.Mol.Biol., 11, 2004
|
|
4BSA
 
 | | Crystal Structure of the Haemagglutinin (with Asn-133 Glycosylation) from an H7N9 Influenza Virus Isolated from Humans | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Xiong, X, Haire, L.F, Martin, S.R, Wharton, S.A, Daniels, R.S, Bennett, M.S, McCauley, J.W, Collins, P.J, Walker, P.A, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor Binding by an H7N9 Influenza Virus from Humans
Nature, 499, 2013
|
|
2B3T
 
 | | Structure of complex between E. coli translation termination factor RF1 and the PrmC methyltransferase | | Descriptor: | Peptide chain release factor 1, Protein methyltransferase hemK, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Graille, M, Heurgue-Hamard, V, Champ, S, Mora, L, Scrima, N, Ulryck, N, van Tilbeurgh, H, Buckingham, R.H. | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for bacterial class I release factor methylation by PrmC
Mol.Cell, 20, 2005
|
|
6X5M
 
 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6GRZ
 
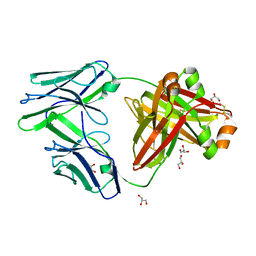 | |
1QCY
 
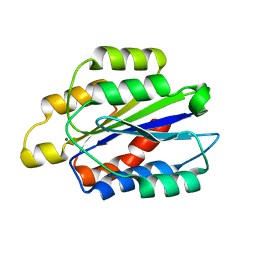 | | THE CRYSTAL STRUCTURE OF THE I-DOMAIN OF HUMAN INTEGRIN ALPHA1BETA1 | | Descriptor: | I-DOMAIN OF INTEGRIN ALPHA1BETA1, MAGNESIUM ION | | Authors: | Kankare, J.A, Salminen, T.A, Nymalm, Y, Kaepylae, J, Heino, J, Johnson, M.S. | | Deposit date: | 1999-05-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Jararhagin-derived RKKH Peptides Induce Structural Changes in a1I Domain of Human Integrin a1b1
J.Biol.Chem., 279, 2004
|
|
2AG1
 
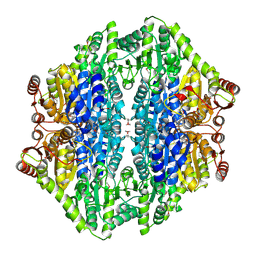 | |
2AH5
 
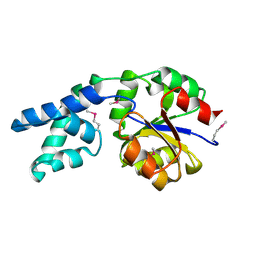 | | Hydrolase, haloacid dehalogenase-like family protein SP0104 from Streptococcus pneumoniae | | Descriptor: | COG0546: Predicted phosphatases | | Authors: | Binkowski, T.A, Zhou, M, Abdullah, J, Collart, F, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Hydrolase, haloacid dehalogenase-like family protein SP0104 from Streptococcus pneumoniae
To be Published
|
|
