8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
1X7B
 
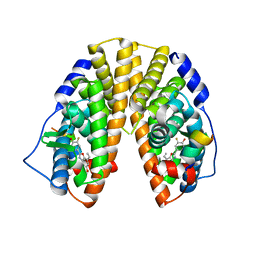 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
7M40
 
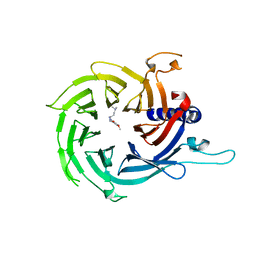 | | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4) | | Descriptor: | Histone-binding protein RBBP4, N~3~-{4-[3-(dimethylamino)pyrrolidin-1-yl]-6,7-dimethoxyquinazolin-2-yl}-N~1~,N~1~-dimethylpropane-1,3-diamine | | Authors: | Perveen, S, Dong, A, Tempel, W, Zepeda-Velazquez, C, Abbey, M, McLeod, D, Marcellus, R, Mohammed, M, Ensan, D, Panagopoulos, D, Trush, V, Gibson, E, Brown, P.J, Arrowsmith, C.H, Schapira, M, Al-awar, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4)
To Be Published
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
7A46
 
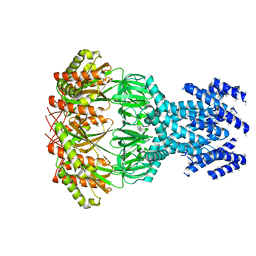 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3SIG
 
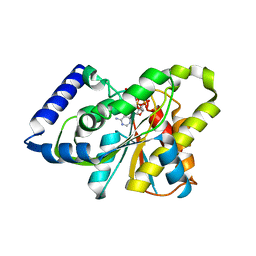 | | The X-ray crystal structure of poly(ADP-ribose) glycohydrolase (PARG) bound to ADP-ribose from Thermomonospora curvata | | Descriptor: | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly(ADP-ribose) glycohydrolase | | Authors: | Leys, D, Dunstan, M.S. | | Deposit date: | 2011-06-18 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase.
Nature, 477, 2011
|
|
7MF0
 
 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS1
 
 | | Structure of SARS-CoV-2 Mpro in complex with nsp12-nsp13 (C12) cut site sequence | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
1XDB
 
 | | Crystal Structure of the Nitrogenase Fe protein Asp129Glu | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
8G46
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
8GNE
 
 | | Crystal structure of human adenosine A2A receptor in complex with an insurmountable inverse agonist, KW-6356. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
8GNG
 
 | | Crystal structure of human adenosine A2A receptor in complex with istradefylline. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 8-[(~{E})-2-(3,4-dimethoxyphenyl)ethenyl]-1,3-diethyl-7-methyl-purine-2,6-dione, Adenosine receptor A2a, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
7AMF
 
 | |
7AMB
 
 | |
2VU5
 
 | | Crystal structure of Pndk from Bacillus anthracis | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Misra, G, Aggarwal, A, Dube, D, Zaman, M.S, Singh, Y, Ramachandran, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bacillus Anthracis Nucleoside Diphosphate Kinase and its Characterization Reveals an Enzyme Adapted to Perform Under Stress Conditions.
Proteins, 76, 2009
|
|
3J8D
 
 | | Cryoelectron microscopy of dengue-Fab E104 complex at pH 5.5 | | Descriptor: | Envelope protein E, antibody E111 Fab fragment, glycoprotein DIII | | Authors: | Zhang, X.Z, Sheng, J, Austin, S.K, Hoornweg, T, Smit, J.M, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2014-10-13 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Structure of Acidic pH Dengue Virus Showing the Fusogenic Glycoprotein Trimers.
J.Virol., 89, 2015
|
|
6BX1
 
 | |
2XYO
 
 | | Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
6ZYD
 
 | | YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI,Low conductance mechanosensitive channel YnaI | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-07-31 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6GRZ
 
 | |
