6HVH
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 1 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(oxan-4-yl)pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Jakubiec, K, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
8Z85
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8YJ7
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, ZINC ION, scFv13 | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
6HVJ
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 3 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, 8-(3-methyl-1-benzofuran-5-yl)-~{N}-(4-methylsulfonylpyridin-3-yl)quinoxalin-6-amine, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
8U37
 
 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8ZNG
 
 | |
8ZNE
 
 | |
8ZNB
 
 | |
8YJ8
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, VHH43, ZINC ION | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8ZNC
 
 | |
8YJ5
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, VHH43 | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8YJ6
 
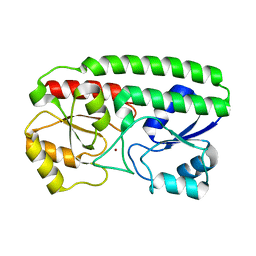 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, ZINC ION | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
8ZND
 
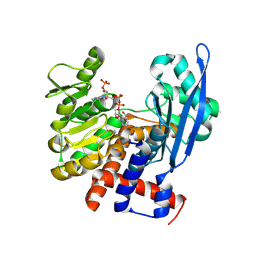 | |
2CHN
 
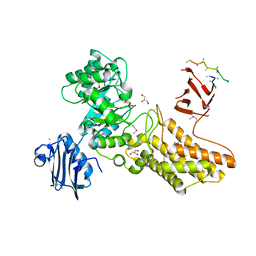 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity- NAG-thiazoline complex | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
8XX0
 
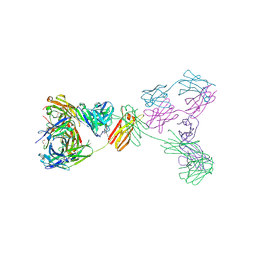 | | Crystal structure of anti-IgE antibody HMK-12 Fab complexed with IgE F(ab')2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SPE7 immunoglobulin E F(ab')2 heavy chain, ... | | Authors: | Hirano, T, Koyanagi, A, Kasai, M, Okumura, K. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric inhibition of IgE-Fc epsilon RI interactions by simultaneous targeting of IgE F(ab')2 epitopes.
Commun Biol, 7, 2024
|
|
7Y6L
 
 | |
7Y6N
 
 | |
2B50
 
 | | Human Nuclear Receptor-Ligand Complex 2 | | Descriptor: | CALCIUM ION, Peroxisome proliferator activated receptor delta, VACCENIC ACID, ... | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
4UV8
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
5DGD
 
 | |
6IC0
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 4 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-pyrimidin-5-yl-pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
2CXV
 
 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
7YC7
 
 | | Dark, fully reduced structure of the MmCPDII-DNA complex as produced at SwissFEL | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD7
 
 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
