7VHD
 
 | | Crystal structure of the STX2a complexed with R4A peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ARG-ARG-ARG-ARG-ALA, Shiga toxin 2 B subunit, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
6ZPR
 
 | | Solution structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein,Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-06 | | Method: | SOLUTION NMR | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A49
 
 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6HVH
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 1 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-(oxan-4-yl)pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Jakubiec, K, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
8P9O
 
 | | PCNA from Chaetomium thermophilum in complex with PolD3 peptide | | Descriptor: | Proliferating cell nuclear antigen, Synthetic peptide corresponding to amino acids 437 to 451 of PolD3 from Chaetomium thermophilum | | Authors: | Alphey, M.S, Wolford, C.B, MacNeill, S.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Canonical binding of Chaetomium thermophilum DNA polymerase delta / zeta subunit PolD3 and flap endonuclease Fen1 to PCNA.
Front Mol Biosci, 10, 2023
|
|
8RHJ
 
 | | Yeast 20S proteasome in complex with a macrocyclic oxindole epoxyketone (compound 5) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Macrocyclic oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
8A5X
 
 | |
6Z9M
 
 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
3KVW
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
7BEG
 
 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7BEF
 
 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7Z6C
 
 | | Crystal structure of human Dihydroorotate Dehydrogenase in complex with the inhibitor 2-Hydroxy-N-(2-ispropyl-5-methyl-4-phenoxyphenyl)pyrazolo[1,5-a]pyridine-3-carboxamide. | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Alberti, M, Lolli, M.L, Boschi, D, Sainas, S, Rizzi, M, Ferraris, D.M, Miggiano, R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting Acute Myelogenous Leukemia Using Potent Human Dihydroorotate Dehydrogenase Inhibitors Based on the 2-Hydroxypyrazolo[1,5- a ]pyridine Scaffold: SAR of the Aryloxyaryl Moiety.
J.Med.Chem., 65, 2022
|
|
8OY0
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Acinetobacter baumanii | | Descriptor: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, GLYCEROL, ... | | Authors: | Alphey, M.S, Read, B, da Silva, R.G. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.
Biochemistry, 63, 2024
|
|
6XKQ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
8CSE
 
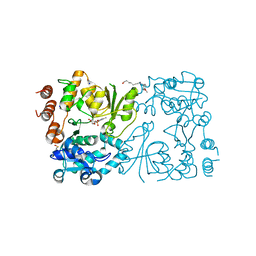 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSD
 
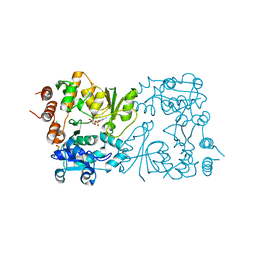 | | WbbB D232C Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSB
 
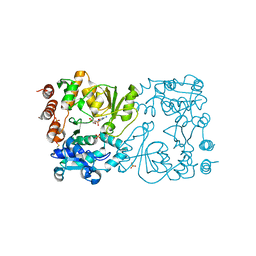 | | WbbB D232N in complex with CMP-beta-Kdo | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSF
 
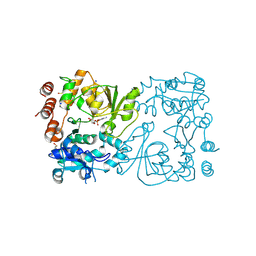 | | WbbB D232C-Kdo adduct + alpha-Rha(1,3)GlcNAc ternary complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSC
 
 | | WbbB D232N-Kdo adduct | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Forrester, T.J.B, Kimber, M.S. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
6ZCI
 
 | | Crystal structure of BRD4-BD1 in complex with NVS-BET-1 | | Descriptor: | (4~{R})-4-(4-chlorophenyl)-1-cyclopropyl-5-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)-3-methyl-4~{H}-pyrrolo[3,4-c]pyrazol-6-one, Bromodomain-containing protein 4 | | Authors: | Faller, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | BET bromodomain inhibitors regulate keratinocyte plasticity.
Nat.Chem.Biol., 17, 2021
|
|
6IBY
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 6 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpyrrolidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6YVU
 
 | | Condensin complex from S.cerevisiae ATP-free apo non-engaged state | | Descriptor: | Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2,Smc2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Aragon, L, Haering, C.H, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8Q7I
 
 | |
6ZBA
 
 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
6ZGM
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with the thiazolecarboxylate inhibitor ANT2681 | | Descriptor: | 5-[[4-(carbamimidamidocarbamoylamino)-3,5-bis(fluoranyl)phenyl]sulfonylamino]-1,3-thiazole-4-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Docquier, J.D, Pozzi, C, Marcoccia, F, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ANT2681: SAR Studies Leading to the Identification of a Metallo-beta-lactamase Inhibitor with Potential for Clinical Use in Combination with Meropenem for the Treatment of Infections Caused by NDM-ProducingEnterobacteriaceae.
Acs Infect Dis., 6, 2020
|
|
