3VKQ
 
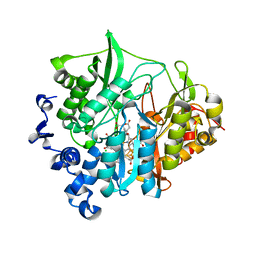 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with middle X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VK4
 
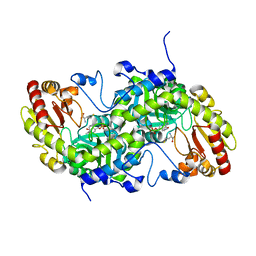 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant complexed with L-homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Methionine gamma-lyase | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
2ZOE
 
 | | HA3 subcomponent of Clostridium botulinum type C progenitor toxin, complex with N-acetylneuramic acid | | Descriptor: | Hemagglutinin components HA3, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Kotani, M, Tonozuka, T, Ide, A, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-05-09 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
2LVA
 
 | | NMR solution structure of the N-terminal domain of human USP28, Northeast structural genomics consortium target HT8470A | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-29 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of human USP28
To be Published
|
|
3VKR
 
 | | Assimilatory nitrite reductase (Nii3) - NO2 complex from tobbaco leaf analysed with high X-ray dose | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, NITRITE ION, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-11-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The reductive reaction mechanism of tobacco nitrite reductase derived from a combination of crystal structures and ultraviolet-visible microspectroscopy
Proteins, 80, 2012
|
|
3VM6
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
1WJ2
 
 | | Solution Structure of the C-terminal WRKY Domain of AtWRKY4 | | Descriptor: | Probable WRKY transcription factor 4, ZINC ION | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis WRKY DNA binding domain.
Plant Cell, 17, 2005
|
|
3VMY
 
 | |
2MHG
 
 | |
2ZPM
 
 | |
3VLZ
 
 | | Assimilatory nitrite reductase (Nii3) - N226K mutant - SO3 full complex from tobacco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray crystal structure of a mutant assimilatory nitrite reductase that shows sulfite reductase-like activity
Chem.Biodivers., 9, 2012
|
|
3VNC
 
 | | Crystal Structure of TIP-alpha N25 from Helicobacter Pylori in its natural dimeric form | | Descriptor: | TIP-alpha | | Authors: | Gao, M, Li, D, Hu, Y, Zou, Q, Wang, D.-C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TNF-alpha-Inducing Protein from Helicobacter Pylori in Active Form Reveals the Intrinsic Molecular Flexibility for Unique DNA-Binding
Plos One, 7, 2012
|
|
2LU6
 
 | | NMR solution structure of Midi peptide designed based on m-conotoxins | | Descriptor: | Midi peptide designed based on m-conotoxins | | Authors: | Dyubankova, N, Lescrinier, E, Stevens, M, Tytgat, J, Herdewijn, P, Peigneur, S. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of bioactive peptides from naturally occurring mu-conotoxin structures.
J.Biol.Chem., 287, 2012
|
|
2M2U
 
 | |
2ZT4
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [810 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4EUH
 
 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | Descriptor: | Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
1EHW
 
 | | HUMAN NUCLEOSIDE DIPHOSPHATE KINASE 4 | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Milon, L, Meyer, P, Chiadmi, M, Munier, A, Johansson, M, Karlsson, A, Lascu, I, Capeau, J, Janin, J, Lacombe, M.-L. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase.
J.Biol.Chem., 275, 2000
|
|
2M57
 
 | |
3VR2
 
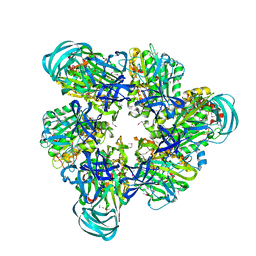 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VS8
 
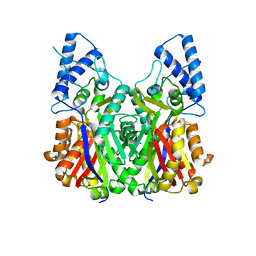 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
378D
 
 | | STRUCTURE OF THE SIDE-BY-SIDE BINDING OF DISTAMYCIN TO DNA | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*GP*TP*AP*TP*AP*TP*AP*C)-3'), SODIUM ION | | Authors: | Mitra, S.N, Wahl, M.C, Sundaralingam, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the side-by-side binding of distamycin to d(GTATATAC)2.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2M0F
 
 | |
3VSP
 
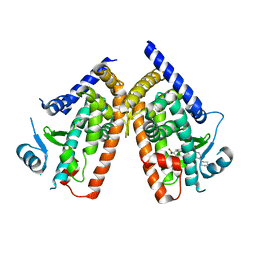 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist mekt28 | | Descriptor: | (2R)-2-benzyl-3-[3-({[4-(piperidin-1-yl)benzoyl]amino}methyl)-4-propoxyphenyl]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Ohashi, M, Morikawa, K, Miyachi, H. | | Deposit date: | 2012-04-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist mekt28
To be Published
|
|
2LW7
 
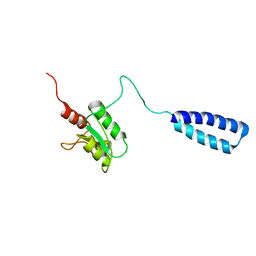 | | NMR solution structure of human HisRS splice variant | | Descriptor: | Histidine--tRNA ligase, cytoplasmic | | Authors: | Ye, F, Wei, Z, Wu, J, Schimmel, P, Zhang, M. | | Deposit date: | 2012-07-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of human HisRS splice variant
To be Published
|
|
2ZXQ
 
 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
