1X40
 
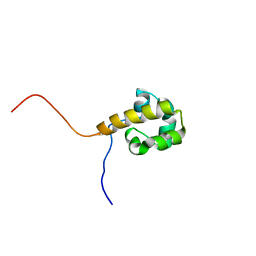 | | Solution structure of the SAM domain of human ARAP2 | | Descriptor: | ARAP2 | | Authors: | Sasagawa, A, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-12 | | Release date: | 2005-11-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM domain of human ARAP2
To be Published
|
|
2RR3
 
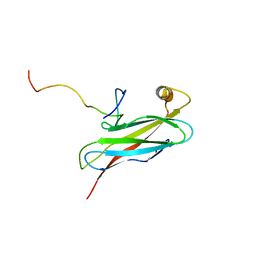 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | Descriptor: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | Authors: | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
3ME1
 
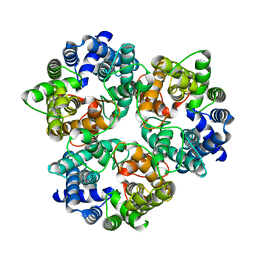 | |
2RS2
 
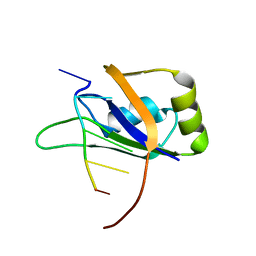 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
3MRW
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina at 1.7 A resolution
To be Published
|
|
1X50
 
 | | Solution structure of the C-terminal gal-bind lectin domain from human galectin-4 | | Descriptor: | Galectin-4 | | Authors: | Tomizawa, T, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal gal-bind lectin domain from human galectin-4
To be Published
|
|
2SN3
 
 | | STRUCTURE OF SCORPION TOXIN VARIANT-3 AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SCORPION NEUROTOXIN (VARIANT 3) | | Authors: | Zhao, B, Carson, M, Ealick, S.E, Bugg, C.E. | | Deposit date: | 1992-02-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of scorpion toxin variant-3 at 1.2 A resolution.
J.Mol.Biol., 227, 1992
|
|
1WGQ
 
 | | Solution Structure of the Pleckstrin Homology Domain of Mouse Ethanol Decreased 4 Protein | | Descriptor: | FYVE, RhoGEF and PH domain containing 6; ethanol decreased 4 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Mouse Ethanol Decreased 4 Protein
To be Published
|
|
3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3LZR
 
 | | Crystal Structure Analysis of Manganese treated P19 protein from Campylobacter jejuni at 2.73 A at pH 9 and Manganese peak wavelength (1.893 A) | | Descriptor: | COPPER (II) ION, MANGANESE (II) ION, P19 protein, ... | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
1WHA
 
 | | Solution structure of the second PDZ domain of human scribble (KIAA0147 protein). | | Descriptor: | KIAA0147 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of human scribble (KIAA0147 protein).
To be Published
|
|
4Z88
 
 | | SH3-II of Drosophila Rim-binding protein with Aplip1 peptide | | Descriptor: | JNK-interacting protein 1, PHOSPHATE ION, RIM-binding protein, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
7ZVJ
 
 | | Homodimeric structure of LARGE1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Diskin, R, Katz, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for matriglycan synthesis by the LARGE1 dual glycosyltransferase.
Plos One, 17, 2022
|
|
3M02
 
 | | The Crystal Structure of 5-epi-aristolochene synthase complexed with (2-cis,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
8A47
 
 | | IdeS in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG-degrading protease, ... | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
4Z8Z
 
 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
4Z98
 
 | | Crystal Structure of Hen Egg White Lysozyme using Serial X-ray Diffraction Data Collection | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Murray, T.D, Lyubimov, A.Y, Ogata, C.M, Uervirojnangkoorn, M, Brunger, A.T, Berger, J.M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3MGU
 
 | | Structure of S. cerevisiae Tpa1 protein, a proline hydroxylase modifying ribosomal protein Rps23 | | Descriptor: | FE (II) ION, PKHD-type hydroxylase TPA1 | | Authors: | Henri, J, Rispal, D, Bayart, E, van Tilbeurgh, H, Seraphin, B, Graille, M. | | Deposit date: | 2010-04-07 | | Release date: | 2010-07-14 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insights into S. cerevisiae Tpa1, a putative prolyl hydroxylase influencing translation termination and transcription
To be Published
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
8A48
 
 | | Less crystallisable" IgG1 Fc fragment (E382S variant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, ... | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.044 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
3NV6
 
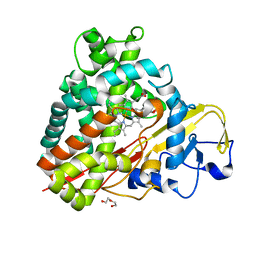 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
1WX4
 
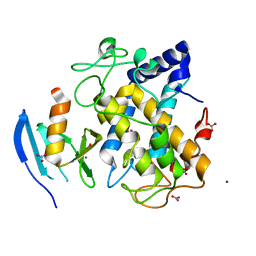 | | Crystal structure of the oxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein prepared by the addition of dithiothreitol | | Descriptor: | COPPER (II) ION, MelC, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
4GB7
 
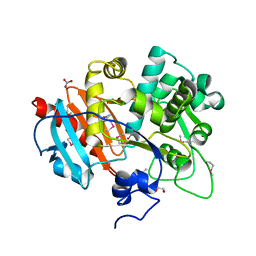 | | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanoate-dimer hydrolase, NITRATE ION | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis.
To be Published
|
|
8A49
 
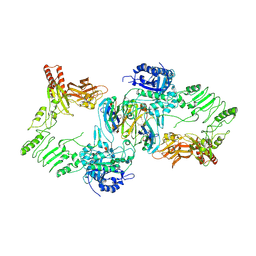 | | Endoglycosidase S in complex with IgG1 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG1 Fc, Secreted endoglycosidase EndoS | | Authors: | Sudol, A.S.L, Tews, I, Crispin, M. | | Deposit date: | 2022-06-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Extensive substrate recognition by the streptococcal antibody-degrading enzymes IdeS and EndoS.
Nat Commun, 13, 2022
|
|
3IHJ
 
 | | Human alanine aminotransferase 2 in complex with PLP | | Descriptor: | Alanine aminotransferase 2, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutamate pyruvate transaminase 2
To be Published
|
|
