6EOU
 
 | |
8D5K
 
 | |
8D5E
 
 | | The complex of Gtf2b Peptide TGAASFDEF Presented by H2-Dd | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Custodio, J.M.F, Baker, B.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and physical features that distinguish tumor-controlling from inactive cancer neoepitopes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D5J
 
 | |
8D5F
 
 | |
4UWN
 
 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
4WN4
 
 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WF1
 
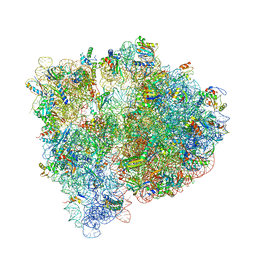 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6F4C
 
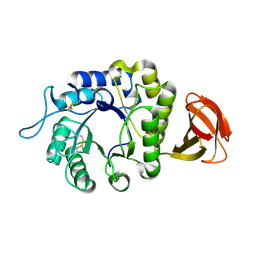 | | Nicotiana benthamiana alpha-galactosidase | | Descriptor: | alpha-galactosidase | | Authors: | Kytidou, K, Aerts, J.M.F.G, Pannu, N.S. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nicotiana benthamianaalpha-galactosidase A1.1 can functionally complement human alpha-galactosidase A deficiency associated with Fabry disease.
J. Biol. Chem., 293, 2018
|
|
4WX2
 
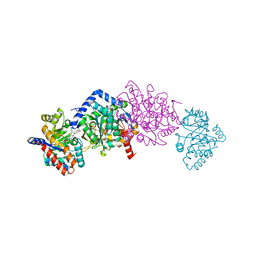 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
6FQI
 
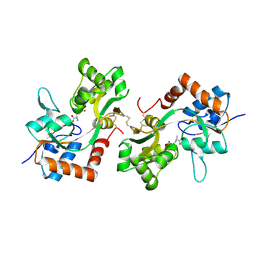 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form B) at 2.91 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.91001153 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
4WSL
 
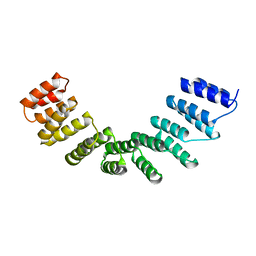 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
6FQK
 
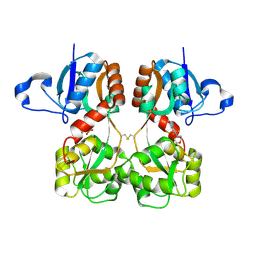 | | GluA2(flop) S729C ligand binding core dimer bound to ZK200775 at 1.98 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.98010445 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6FQJ
 
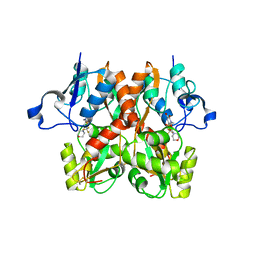 | | GluA2(flop) G724C ligand binding core dimer bound to ZK200775 at 2.50 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.500017 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
8DSO
 
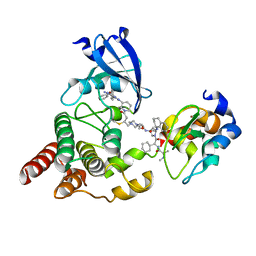 | | Structure of cIAP1, BTK and BCCov | | Descriptor: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide bound form, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | Authors: | Schiemer, J.S, Calabrese, M.F. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
6FQG
 
 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form A) at 2.34 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34139085 Å) | | Cite: | X-ray structure of GluA2 flop G724C ligand binding core dimer bound to glutamate at 2.32 Angstroms resolution
To Be Published
|
|
8DSF
 
 | | Structure of cIAP1 with BCCov | | Descriptor: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide unbound form, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Schiemer, J.S, Calabrese, M.F. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
4X3V
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
6FQH
 
 | | GluA2(flop) S729C ligand binding core dimer bound to NBQX at 1.76 Angstrom resolution | | Descriptor: | 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75940013 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
8CP7
 
 | |
6XE3
 
 | | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site. | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-06-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Salmonella typhimurium Tryptophan Synthase beta-S377A mutant in complex with inhibitor F9 at the enzyme alpha-site, cesium ion at the metal coordination site and carbanion III E(C3) at the enzyme beta-site.
To be Published
|
|
6XIN
 
 | | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site. | | Descriptor: | (2R,3Z)-3-amino-3-imino-2-[(E)-phenyldiazenyl]propanamide, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Chang, C, Mueller, L.J, Dunn, M.F, Fan, L. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site.
To Be Published
|
|
6XSY
 
 | | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Mueller, L.J, Dunn, M.F. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site.
To be Published
|
|
6Y3Y
 
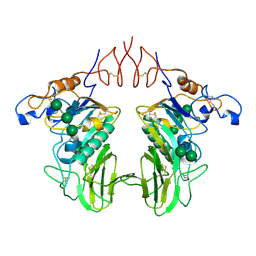 | | Human Coronavirus HKU1 Haemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Hurdiss, D.L, Drulyte, I, Pronker, M.F. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans.
Nat Commun, 11, 2020
|
|
