5LMR
 
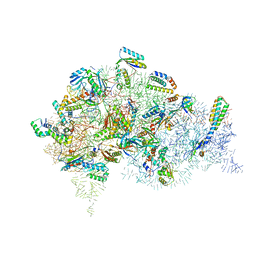 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMO
 
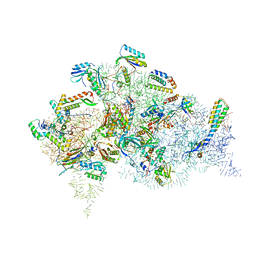 | | Structure of bacterial 30S-IF1-IF3-mRNA translation pre-initiation complex (state-1B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMV
 
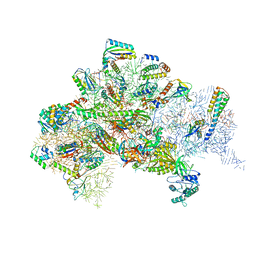 | | Structure of bacterial 30S-IF1-IF2-IF3-mRNA-tRNA translation pre-initiation complex(state-III) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMP
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA translation pre-initiation complex (state-1C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.35 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
8P9Y
 
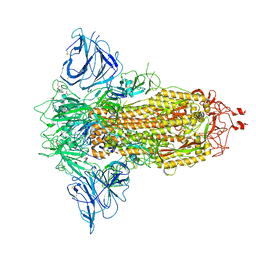 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
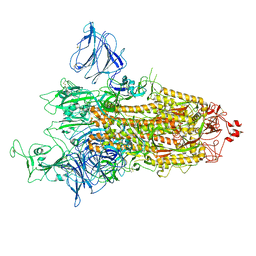 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8S8K
 
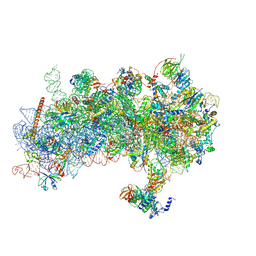 | | Structure of a yeast 48S-AUC preinitiation complex in swivelled conformation (model py48S-AUC-swiv-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S12, 40S ribosomal protein S21, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
8S8H
 
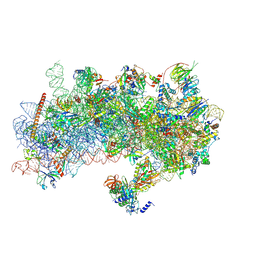 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8D
 
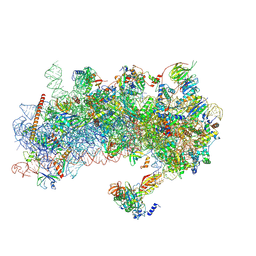 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8G
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8RW1
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8I
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
3J80
 
 | | CryoEM structure of 40S-eIF1-eIF1A preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
3J81
 
 | | CryoEM structure of a partial yeast 48S preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-29 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
4OZL
 
 | | GlnK2 from Haloferax mediterranei complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Palanca, C, Pedro-Roig, L, Llacer, J.L, Camacho, M, Bonete, M.J, Rubio, V. | | Deposit date: | 2014-02-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4942 Å) | | Cite: | The structure of a PII signaling protein from a halophilic archaeon reveals novel traits and high-salt adaptations.
Febs J., 281, 2014
|
|
4OZN
 
 | | GlnK2 from Haloferax mediterranei complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Palanca, C, Pedro-Roig, L, Llacer, J.L, Camacho, M, Bonete, M.J, Rubio, V. | | Deposit date: | 2014-02-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a PII signaling protein from a halophilic archaeon reveals novel traits and high-salt adaptations.
Febs J., 281, 2014
|
|
4OZJ
 
 | | GlnK2 from Haloferax mediterranei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Palanca, C, Pedro-Roig, L, Llacer, J.L, Camacho, M, Bonete, M.J, Rubio, V. | | Deposit date: | 2014-02-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a PII signaling protein from a halophilic archaeon reveals novel traits and high-salt adaptations.
Febs J., 281, 2014
|
|
2JER
 
 | |
