1NXC
 
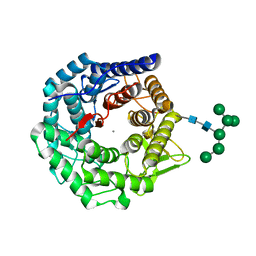 | | Structure of mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) | | Descriptor: | CALCIUM ION, Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Tempel, W, Liu, Z.-J, Karaveg, K, Rose, J, Moremen, K.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-02-10 | | Release date: | 2004-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of mouse Golgi alpha-mannosidase IA reveals the molecular basis for substrate specificity among class 1 (family 47 glycosylhydrolase) alpha1,2-mannosidases
J.Biol.Chem., 279, 2004
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2B
 
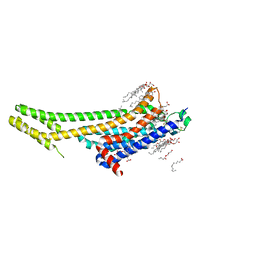 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2A
 
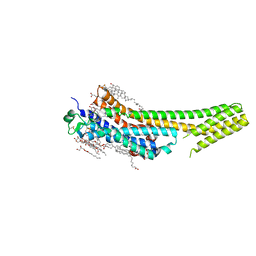 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2C
 
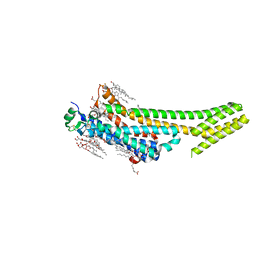 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
4O8U
 
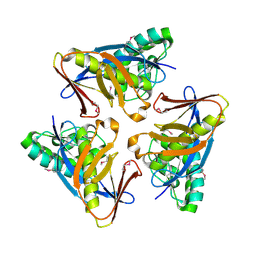 | | Structure of PF2046 | | Descriptor: | Uncharacterized protein PF2046 | | Authors: | Su, J, Liu, Z.-J. | | Deposit date: | 2013-12-30 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Crystal structure of a novel non-Pfam protein PF2046 solved using low resolution B-factor sharpening and multi-crystal averaging methods
Protein Cell, 1, 2010
|
|
3HY4
 
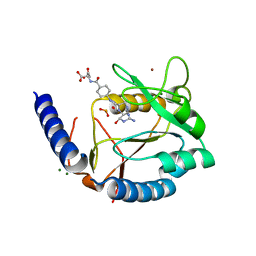 | | Structure of human MTHFS with N5-iminium phosphate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({trans-4-[({(2R,4R,4aS,6S,8aS)-2-amino-4-hydroxy-5-[(phosphonooxy)methyl]decahydropteridin-6-yl}methyl)amino]cyclohexyl}carbonyl)-L-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY6
 
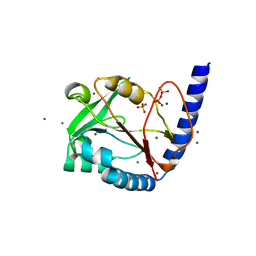 | | Structure of human MTHFS with ADP | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HY3
 
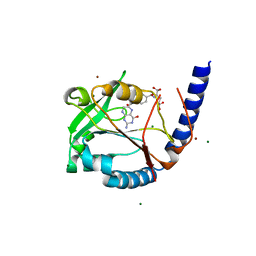 | | Structure of human MTHFS with 10-formyltetrahydrofolate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({4-[{[(2R,4S,4aR,6S,8aS)-2-amino-4-hydroxydecahydropteridin-6-yl]methyl}(formyl)amino]phenyl}carbonyl)-D-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
3HXT
 
 | | Structure of human MTHFS | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Zhang, R, Joachimiak, A, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
5UVM
 
 | | HIT family hydrolase from Clostridium thermocellum Cth-393 | | Descriptor: | ADENOSINE, Histidine triad (HIT) protein, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J, Tempel, W, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIT family hydrolase from Clostridium thermocellum Cth-393
To Be Published
|
|
5F9P
 
 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
3UX9
 
 | | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus | | Descriptor: | Interferon alpha-1/13, ScFv antibody | | Authors: | Ouyang, S, Zhao, L.X, Liang, W, Shaw, N, Liu, Z.-J, Liang, M.-F. | | Deposit date: | 2011-12-04 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus
J.Mol.Med., 2012
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
4JBM
 
 | | Structure of murine DNA binding protein bound with ds DNA | | Descriptor: | DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), Interferon-inducible protein AIM2 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBJ
 
 | | Structural mimicry for functional antagonism | | Descriptor: | Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Ma, F, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
4JBK
 
 | | Molecular basis for abrogation of activation of pro-inflammatory cytokines | | Descriptor: | DNA (5'-D(P*GP*GP*AP*AP*TP*TP*AP*TP*AP*AP*TP*TP*CP*C)-3'), Interferon-activable protein 202 | | Authors: | Ru, H, Ni, X, Crowley, C, Zhao, L, Ding, W, Hung, L.-W, Shaw, N, Cheng, G, Liu, Z.-J. | | Deposit date: | 2013-02-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Structural basis for termination of AIM2-mediated signaling by p202
Cell Res., 23, 2013
|
|
5BPJ
 
 | | All Three Ca(2+)-binding Loops of Light-sensitive Ctenophore Photoprotein Berovin Bind Magnesium Ions: The Spatial Structure of Mg(2+)-loaded Apo-berovin | | Descriptor: | Apoberovin 1, MAGNESIUM ION | | Authors: | Burakova, L.P, Natashin, P.V, Malikova, N.P, Niu, F, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2015-05-28 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | All Ca(2+)-binding loops of light-sensitive ctenophore photoprotein berovin bind magnesium ions: The spatial structure of Mg(2+)-loaded apo-berovin.
J. Photochem. Photobiol. B, Biol., 154, 2016
|
|
4DKS
 
 | | A spindle-shaped virus protein (chymotrypsin treated) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-02-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2HJM
 
 | |
1S30
 
 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | Deposit date: | 2004-01-12 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
1S2Z
 
 | | X-ray crystal structure of Desulfovibrio vulgaris Rubrerythrin with displacement of iron by zinc at the diiron Site | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.-J, Rose, J, Wang, B.-C. | | Deposit date: | 2004-01-12 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Displacement of iron by zinc at the diiron site of Desulfovibrio vulgaris rubrerythrin: X-ray crystal structure and anomalous scattering analysis
J.Inorg.Biochem., 98, 2004
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
