8HOM
 
 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-12-10 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir
To Be Published
|
|
8HOL
 
 | |
8HOZ
 
 | |
5WFU
 
 | |
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | 著者 | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-05-11 | | 公開日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
8HCZ
 
 | |
8HDF
 
 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
5X62
 
 | |
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | 著者 | Wang, H, Liu, X, Sun, L, Yang, H. | | 登録日 | 2022-11-03 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7EEY
 
 | |
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8IOC
 
 | | Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | 著者 | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S. | | 登録日 | 2023-03-10 | | 公開日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
6IJZ
 
 | | Structure of a plant cation channel | | 分子名称: | Calcium permeable stress-gated cation channel 1 | | 著者 | Sun, L, Wang, J, Liu, X. | | 登録日 | 2018-10-12 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
5Y3U
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia) | | 分子名称: | PSTD-22AA-PolySia | | 著者 | Liao, S.M, Liu, X.H, Lu, B, Peng, L.X, Chen, D, Huang, R.B, Zhou, G.P. | | 登録日 | 2017-07-31 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia)
To Be Published
|
|
8INQ
 
 | |
8INT
 
 | |
8INW
 
 | |
8INY
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir | | 分子名称: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M, Liu, X. | | 登録日 | 2023-03-10 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir
To Be Published
|
|
8INU
 
 | |
8INX
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir | | 分子名称: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M, Liu, X. | | 登録日 | 2023-03-10 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir
To Be Published
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | 分子名称: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | 著者 | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | 登録日 | 2022-03-02 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7VP8
 
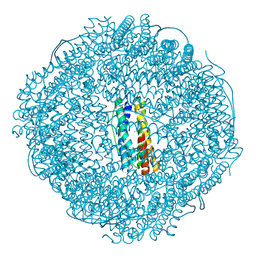 | | Crystal structure of ferritin from Ureaplasma urealyticum | | 分子名称: | CHLORIDE ION, FE (III) ION, Ferritin-like diiron domain-containing protein | | 著者 | Wang, W, Liu, X, Wang, Y, Fu, D, Wang, H. | | 登録日 | 2021-10-15 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Distinct structural characteristics define a new subfamily of Mycoplasma ferritin
Chin.Chem.Lett., 33, 2022
|
|
5JLY
 
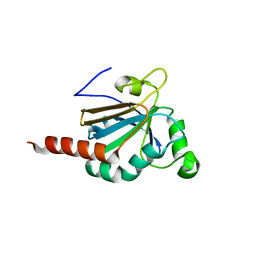 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | 分子名称: | Thioredoxin peroxidase-1 | | 著者 | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | 登録日 | 2016-04-27 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.051 Å) | | 主引用文献 | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
