8ISC
 
 | | Crystal structure of MV in complex with LLP | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of MV in complex with LLP
To Be Published
|
|
8IOZ
 
 | | Crystal structure of transaminase | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | structure of aminotransferase
To Be Published
|
|
8J5N
 
 | |
8YAU
 
 | |
8YAI
 
 | | Crystal structure of glucose 1-dehydrogenase mutant1 from Limosilactobacillus fermentum | | Descriptor: | SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
8YAV
 
 | | Crystal structure of glucose 1-dehydrogenase from Limosilactobacillus fermentum | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-10 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir.
Adv.Synth.Catal., n/a, 2024
|
|
8ZAX
 
 | | Crystal structure of a short-chain dehydrogenase from Lactobacillus fermentum with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SDR family oxidoreductase | | Authors: | Wang, J.J, Cong, L, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-04-25 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
1JYR
 
 | | Xray Structure of Grb2 SH2 Domain Complexed with a Phosphorylated Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, peptide: PSpYVNVQN | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
1JYQ
 
 | | Xray Structure of Grb2 SH2 Domain Complexed with a Highly Affine Phospho Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, mAZ-pY-(alpha Me)pY-N-NH2 peptide inhibitor | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
1JYU
 
 | | Xray Structure of Grb2 SH2 Domain | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
6XRE
 
 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
7BPC
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BP1
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
5IE7
 
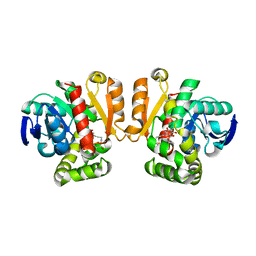 | | Crystal structure of a lactonase double mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE6
 
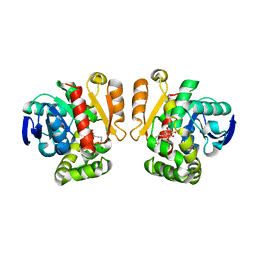 | | Crystal structure of a lactonase mutant in complex with substrate b | | Descriptor: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE4
 
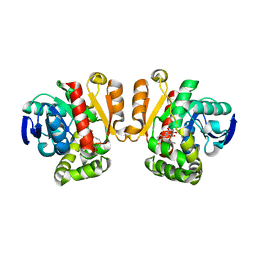 | | Crystal structure of a lactonase mutant in complex with substrate a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE5
 
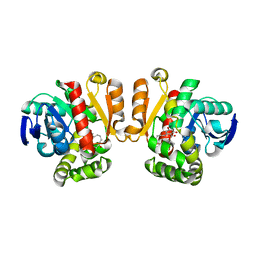 | | Crystal structure of a lactonase double mutant in complex with substrate a | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | Authors: | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-25 | | Release date: | 2017-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
7XG6
 
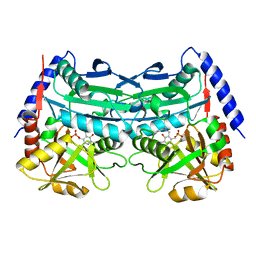 | | Crystal structure of an (R)-selective omega-transaminase mutant from Aspergillus terreus with covalently bound PLP | | Descriptor: | omega-transaminase | | Authors: | Xiang, C, Wu, S.K, Weber, G, Liu, W.D, Wei, R, Bornscheuer, U.T. | | Deposit date: | 2022-04-03 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A growth selection system for the directed evolution of amine-forming or converting enzymes.
Nat Commun, 13, 2022
|
|
7XG5
 
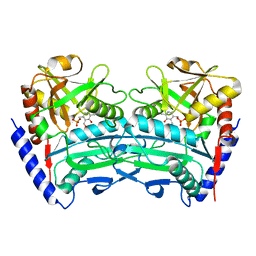 | | Crystal structure of an (R)-selective omega-transaminase mutant from Aspergillus terreus with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, omega-transaminase | | Authors: | Xiang, C, Wu, S.K, Weber, G, Liu, W.D, Wei, R, Bornscheuer, U.T. | | Deposit date: | 2022-04-03 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A growth selection system for the directed evolution of amine-forming or converting enzymes.
Nat Commun, 13, 2022
|
|
7XKS
 
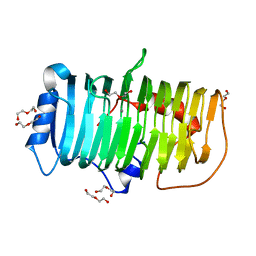 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
4RWA
 
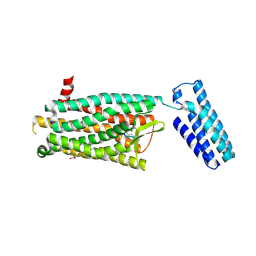 | | Synchrotron structure of the human delta opioid receptor in complex with a bifunctional peptide (PSI community target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Soluble cytochrome b562,Delta-type opioid receptor, bifunctional peptide | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RWD
 
 | | XFEL structure of the human delta opioid receptor in complex with a bifunctional peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, SODIUM ION, ... | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7JPY
 
 | |
