7TQ7
 
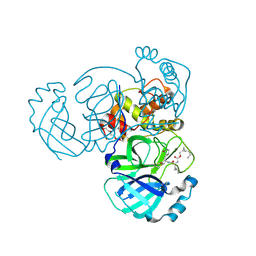 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 13c | | Descriptor: | N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucinamide, Orf1a protein, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4FLN
 
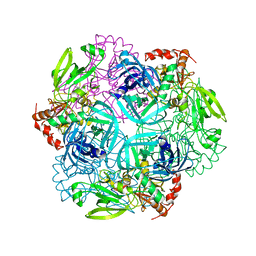 | | Crystal structure of plant protease Deg2 | | Descriptor: | Protease Do-like 2, chloroplastic, Unknown peptide | | Authors: | Gong, W, Liu, L, Sun, R, Gao, F. | | Deposit date: | 2012-06-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
6BTA
 
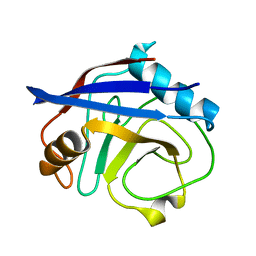 | | CypA Mutant - S99T C115S | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Fraser, J.S, Kenner, L.R, Liu, L. | | Deposit date: | 2017-12-06 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescue of conformational dynamics in enzyme catalysis by directed evolution.
Nat Commun, 9, 2018
|
|
7Y17
 
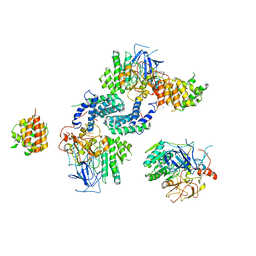 | |
7Y16
 
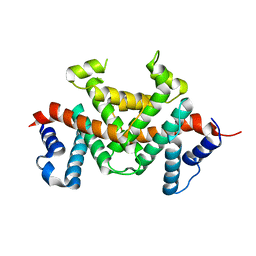 | | Crystal structure of rRNA-processing protein Las1 | | Descriptor: | LAS1 protein | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
7Y18
 
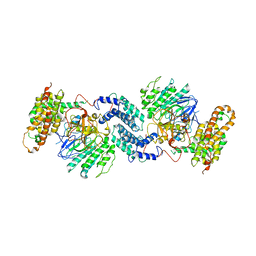 | |
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
8U5O
 
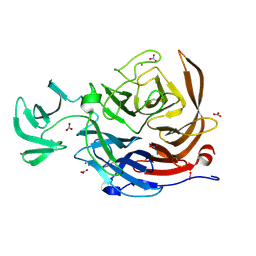 | | The structure of the catalytic domain of NanI sialdase in complex with Neu5Gc | | Descriptor: | CALCIUM ION, Exo-alpha-sialidase, N-glycolyl-alpha-neuraminic acid, ... | | Authors: | Medley, B.J, Low, K.E, Garber, J.M, Gray, T.E, Liu, L, Klassen, L, Fordwour, O.B, Inglis, G.D, Boons, G.J, Zandberg, W.F, Abbott, W.D, Boraston, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens.
J.Biol.Chem., 300, 2024
|
|
7JY5
 
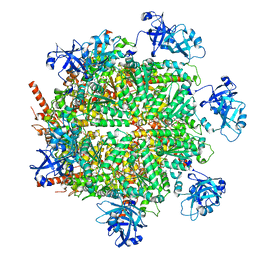 | | Structure of human p97 in complex with ATPgammaS and Npl4/Ufd1 (masked around p97) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative.
Nat Commun, 12, 2021
|
|
7KEO
 
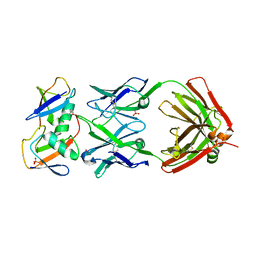 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7JH6
 
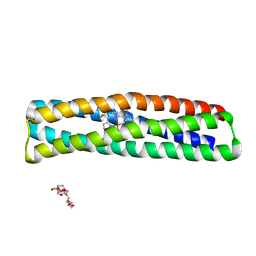 | | De novo designed two-domain di-Zn(II) and porphyrin-binding protein | | Descriptor: | NONAETHYLENE GLYCOL, Two-domain di-Zn(II) and porphyrin-binding protein, ZINC ION, ... | | Authors: | Schmidt, N, Liu, L, DeGrado, W.F. | | Deposit date: | 2020-07-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Allosteric cooperation in a de novo-designed two-domain protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GRQ
 
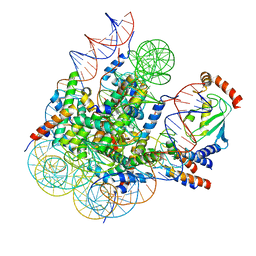 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GRM
 
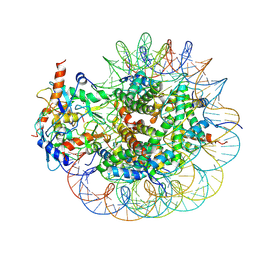 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
7ZBT
 
 | | Subtomogram averaging of Rubisco from native Halothiobacillus carboxysomes | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small subunit | | Authors: | Ni, T, Zhu, Y, Yu, X, Sun, Y, Liu, L, Zhang, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
7LN1
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN3
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMZ
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMY
 
 | | Cryo-EM structure of human p97 in complex with NMS-873 in the presence of ATP, Npl4/Ufd1, and Ub6 | | Descriptor: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN2
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN0
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN4
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN6
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 2, Open State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN5
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 1, Close State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
