6MQ2
 
 | |
7KEO
 
 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7JY5
 
 | | Structure of human p97 in complex with ATPgammaS and Npl4/Ufd1 (masked around p97) | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative.
Nat Commun, 12, 2021
|
|
5YEI
 
 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
7LMZ
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN4
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMY
 
 | | Cryo-EM structure of human p97 in complex with NMS-873 in the presence of ATP, Npl4/Ufd1, and Ub6 | | Descriptor: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN2
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN0
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN6
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 2, Open State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN5
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 1, Close State) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN1
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN3
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pan, M, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5Z9X
 
 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5Z9Z
 
 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
3PS5
 
 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GM8
 
 | | Crystal structure of human WD repeat domain 5 with compound MM-102 | | Descriptor: | MM-102, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
8I6Y
 
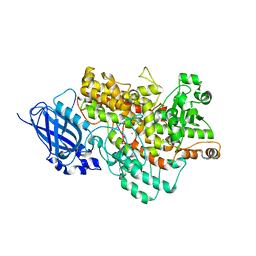 | | Crystal structure of Arabidopsis thaliana LOX1 | | Descriptor: | FE (III) ION, Linoleate 9S-lipoxygenase 1 | | Authors: | Liu, X, Liu, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | UV-B light signal mediates stomatal closure by activating the 9-lipoxygenase pathway
To Be Published
|
|
4IC6
 
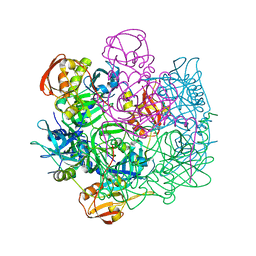 | | Crystal structure of Deg8 | | Descriptor: | Protease Do-like 8, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IC5
 
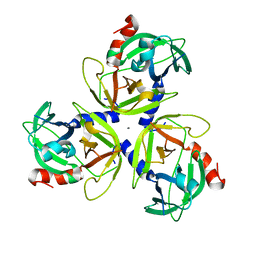 | | Crystal structure of Deg5 | | Descriptor: | CALCIUM ION, Protease Do-like 5, chloroplastic | | Authors: | Gong, W, Sun, W, Fan, H, Gao, F, Liu, L. | | Deposit date: | 2012-12-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | The structures of Arabidopsis Deg5 and Deg8 reveal new insights into HtrA proteases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J4G
 
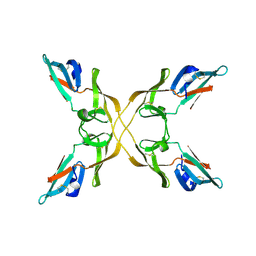 | |
4J4F
 
 | |
4J4E
 
 | |
