7D60
 
 | | Cryo-EM Structure of human CALHM5 in the presence of rubidium red | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7F5X
 
 | | GK domain of Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-L-GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5V
 
 | | Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5U
 
 | |
7V8E
 
 | | Crystal structure of IpaH1.4 LRR domain bound to HOIL-1L UBL domain. | | Descriptor: | RING-type E3 ubiquitin transferase, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8G
 
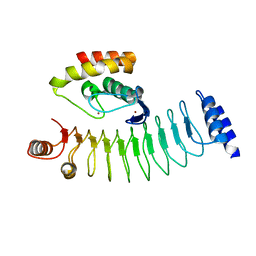 | | Crystal structure of HOIP RING1 domain bound to IpaH1.4 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, RING-type E3 ubiquitin transferase, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8F
 
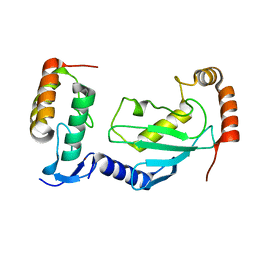 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8H
 
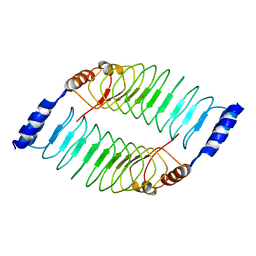 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E35
 
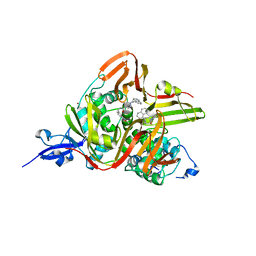 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D61
 
 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D6H
 
 | |
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
1HUO
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*C)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
1HUZ
 
 | | CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP | | Descriptor: | 5'-D(*AP*AP*TP*AP*GP*GP*CP*GP*TP*CP*G)-3', 5'-D(P*CP*GP*AP*CP*GP*CP*CP*T)-3', CHROMIUM ION, ... | | Authors: | Arndt, J.W, Gong, W, Zhong, X, Showalter, A.K, Liu, J, Lin, Z, Paxson, C, Tsai, M.-D, Chan, M.K. | | Deposit date: | 2001-01-04 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes.
Biochemistry, 40, 2001
|
|
3KBG
 
 | | Crystal structure of the 30S ribosomal protein S4e from Thermoplasma acidophilum. Northeast Structural Genomics Consortium Target TaR28. | | Descriptor: | 30S ribosomal protein S4e | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Belote, R.L, Ciccosanti, C, Patel, D, Liu, J, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the 30S ribosomal protein S4e from Thermoplasma acidophilum.
To be Published
|
|
3KLW
 
 | | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132. | | Descriptor: | Primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Vorobiev, S.M, Seetharaman, J, Forouhar, F, Wang, H, Janjua, H, Maglaqui, M, Xiao, T, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132.
To be Published
|
|
3JC6
 
 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5WWU
 
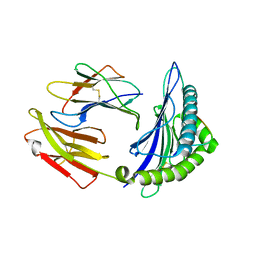 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5WXC
 
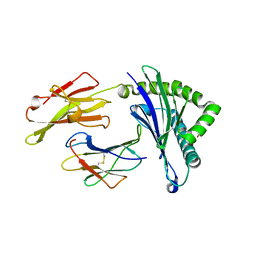 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 2) | | Descriptor: | Beta-2-microglobulin, H7-25-F, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
2PSB
 
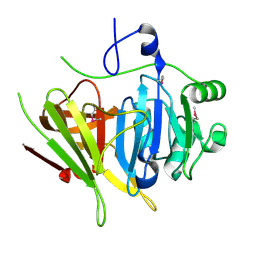 | | Crystal structure of YerB protein from Bacillus subtilis. NorthEast Structural Genomics target SR586 | | Descriptor: | YerB protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-04 | | Release date: | 2007-05-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YerB protein from Bacillus subtilis.
To be Published
|
|
3C86
 
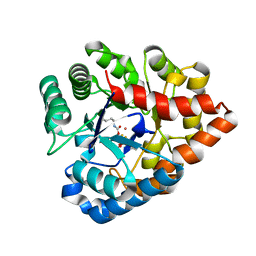 | | OpdA from agrobacterium radiobacter with bound product diethyl thiophosphate from crystal soaking with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2008-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
