1VF4
 
 | | cGSTA1-1 apo form | | Descriptor: | ACETIC ACID, Glutathione S-transferase 3 | | Authors: | Lin, S.C, Lo, Y.C, Tam, M.F, Liaw, Y.C. | | Deposit date: | 2004-04-08 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of chicken glutathione S-transferase A1-1
To be Published
|
|
3DKB
 
 | | Crystal Structure of A20, 2.5 angstrom | | Descriptor: | Tumor necrosis factor, alpha-induced protein 3 | | Authors: | Lin, S.-C, Chung, J.Y, Lo, Y.-C, Wu, H. | | Deposit date: | 2008-06-24 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20
J.Mol.Biol., 376, 2008
|
|
3OIE
 
 | | Crystal structure of the DB1880-D(CGCGAATTCGCG)2 complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, N',N''-{furan-2,5-diylbis[3-(piperidin-4-yloxy)benzene-4,1-diyl]}dipyridine-2-carboximidamide | | Authors: | Lin, S, Neidle, S, Campbell, N. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DB1880-D(CGCGAATTCGCG)2 complex
To be Published
|
|
1VF2
 
 | |
1VF3
 
 | | cGSTA1-1 in complex with glutathione conjugate of CDNB | | Descriptor: | ACETIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase 3 | | Authors: | Lin, S.C, Lo, Y.C, Tam, M.F, Liaw, Y.C. | | Deposit date: | 2004-04-07 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of chicken glutathione S-transferase A1-1
To be Published
|
|
4KXJ
 
 | | Crystal structure of HCoV-OC43 N-NTD complexed with PJ34 | | Descriptor: | Nucleoprotein, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Lin, S.Y, Hou, M.H. | | Deposit date: | 2013-05-27 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target
J.Med.Chem., 57, 2014
|
|
8XR4
 
 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR3
 
 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
4LI4
 
 | |
4LM9
 
 | |
4LMC
 
 | |
4LM7
 
 | |
2POI
 
 | | Crystal structure of XIAP BIR1 domain (I222 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Lin, S. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
3QW9
 
 | | Crystal structure of betaglycan ZP-C domain | | Descriptor: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lin, S.J, Jardetzky, T.S. | | Deposit date: | 2011-02-27 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1PP6
 
 | |
2POM
 
 | | TAB1 with manganese ion | | Descriptor: | MANGANESE (II) ION, Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 | | Authors: | Lin, S.C. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
2POP
 
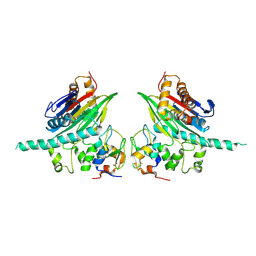 | | The Crystal Structure of TAB1 and BIR1 complex | | Descriptor: | Baculoviral IAP repeat-containing protein 4, Mitogen-activated protein kinase kinase kinase 7-interacting protein 1, ZINC ION | | Authors: | Lin, S.C. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
1PP0
 
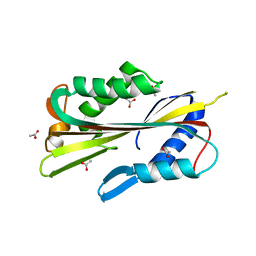 | | volvatoxin A2 in monoclinic crystal | | Descriptor: | ACETIC ACID, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7E9G
 
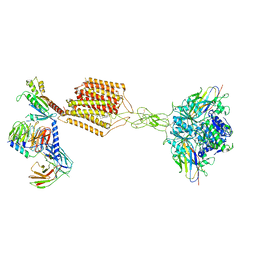 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E9H
 
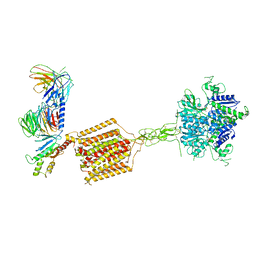 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
1VF1
 
 | |
2QRA
 
 | | Crystal structure of XIAP BIR1 domain (P21 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ETHANOL, ZINC ION | | Authors: | Lin, S.-C. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the BIR1 domain of XIAP in two crystal forms
J.Mol.Biol., 372, 2007
|
|
