5HH3
 
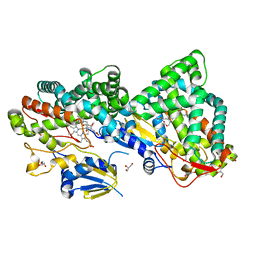 | | OxyA from Actinoplanes teichomyceticus | | Descriptor: | ACETATE ION, GLYCEROL, OxyA protein, ... | | Authors: | Haslinger, K, Cryle, M.J. | | Deposit date: | 2016-01-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of OxyAtei : completing our picture of the glycopeptide antibiotic producing Cytochrome P450 cascade.
Febs Lett., 590, 2016
|
|
1YGP
 
 | | PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE. | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, YEAST GLYCOGEN PHOSPHORYLASE | | Authors: | Lin, K, Rath, V.L, Dai, S.C, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A protein phosphorylation switch at the conserved allosteric site in GP.
Science, 273, 1996
|
|
4TVF
 
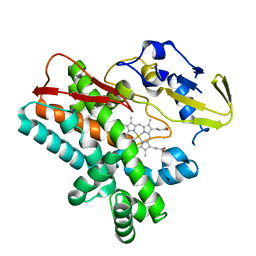 | | OxyB from Actinoplanes teichomyceticus | | Descriptor: | OxyB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytochrome P450 OxyBtei Catalyzes the First Phenolic Coupling Step in Teicoplanin Biosynthesis.
Chembiochem, 15, 2014
|
|
2L1X
 
 | |
6OCK
 
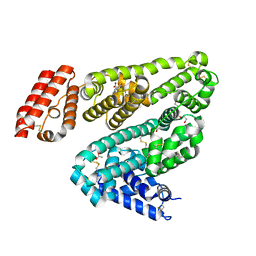
 | | Crystal Structure of Leporine Serum Albumin in Complex with Ketoprofen | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, ... | | Authors: | Zielinski, K, Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
6OCL
 
 | | Crystal Structure of Leporine Serum Albumin in Complex with Suprofen | | Descriptor: | (2S)-2-[4-(thiophene-2-carbonyl)phenyl]propanoic acid, ACETATE ION, Serum albumin, ... | | Authors: | Zielinski, K, Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
7PP4
 
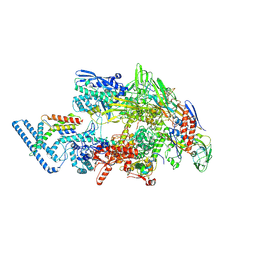 | |
7Q59
 
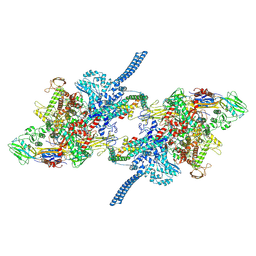 | |
7Q4U
 
 | |
3I3L
 
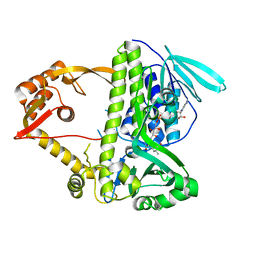 | | Crystal structure of CmlS, a flavin-dependent halogenase | | Descriptor: | Alkylhalidase CmlS, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Podzelinska, K, Soares, A, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-06-30 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chloramphenicol Biosynthesis: The Structure of CmlS, a Flavin-Dependent Halogenase Showing a Covalent Flavin-Aspartate Bond
J.Mol.Biol., 397, 2010
|
|
3P2U
 
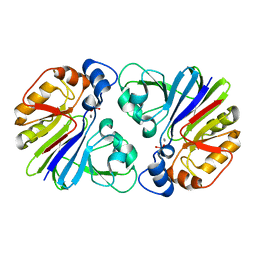 | | Crystal structure of PhnP in complex with orthovanadate | | Descriptor: | MANGANESE (II) ION, Protein phnP, VANADATE ION, ... | | Authors: | Podzelinska, K, Kim, Y.M, Zechel, D, Faucher, F, Jia, Z. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of PhnP in complex with orthovanadate
TO BE PUBLISHED
|
|
7Z8Q
 
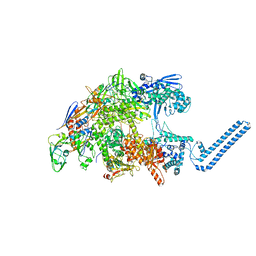 | | Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Brodolin, K. | | Deposit date: | 2022-03-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization
Nat Commun, 14, 2023
|
|
6UL2
 
 | |
4OR0
 
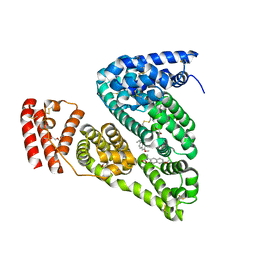 | | Crystal Structure of Bovine Serum Albumin in complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Zielinski, K, Bujacz, A, Sekula, B, Bujacz, G. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen.
Proteins, 82, 2014
|
|
4IC2
 
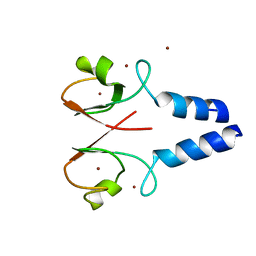 | | Crystal structure of the XIAP RING domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, NICKEL (II) ION, ZINC ION | | Authors: | Linke, K, Nakatani, Y, Day, C.L. | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase
Biochem.J., 450, 2013
|
|
4JK4
 
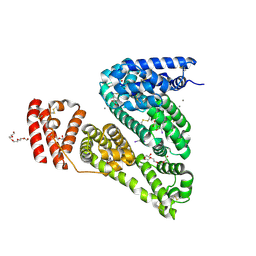 | | Crystal Structure of Bovine Serum Albumin in complex with 3,5-diiodosalicylic acid | | Descriptor: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zielinski, K, Bujacz, A, Sekula, B, Bujacz, G. | | Deposit date: | 2013-03-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Crystallographic studies of the complexes of bovine and equine serum albumin with 3,5-diiodosalicylic acid.
Int.J.Biol.Macromol., 60C, 2013
|
|
2MR8
 
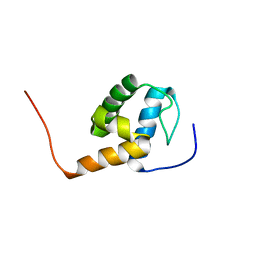 | |
4PXH
 
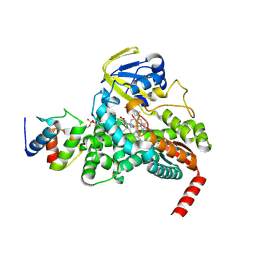 | |
4PWV
 
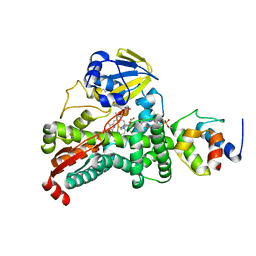 | |
4BY7
 
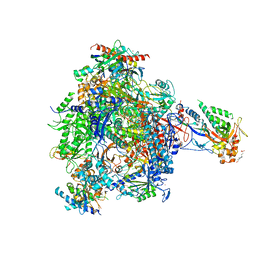 | | elongating RNA Polymerase II-Bye1 TLD complex | | Descriptor: | , 5'-D(*DAP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*DTP)-3', 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
2MR7
 
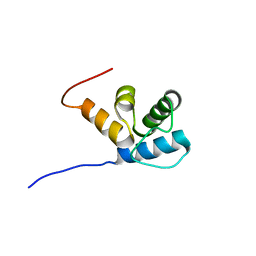 | |
3G1P
 
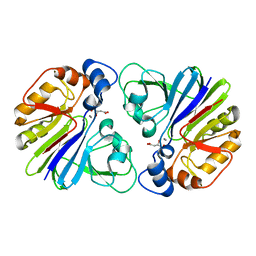 | |
4BXZ
 
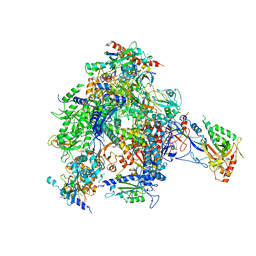 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BY1
 
 | | elongating RNA Polymerase II-Bye1 TLD complex soaked with AMPCPP | | Descriptor: | 5'-D(*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP *TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*AP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4PO0
 
 | | Crystal Structure of Leporine Serum Albumin in complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Serum albumin | | Authors: | Zielinski, K, Bujacz, A, Sekula, B, Bujacz, G. | | Deposit date: | 2014-02-23 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen.
Proteins, 82, 2014
|
|
