2MBY
 
 | | NMR Structure of Rrp7 C-terminal Domain | | Descriptor: | Ribosomal RNA-processing protein 7 | | Authors: | Lin, J, Feng, Y, Ye, K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA-Binding Complex Involved in Ribosome Biogenesis Contains a Protein with Homology to tRNA CCA-Adding Enzyme.
Plos Biol., 11, 2013
|
|
4E08
 
 | |
2QD0
 
 | | Crystal structure of mitoNEET | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Lin, J, Zhou, T, Ye, K, Wang, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6NAD
 
 | |
6VSW
 
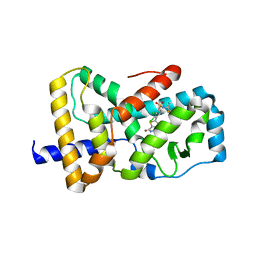 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8GDO
 
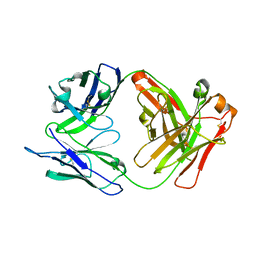 | |
1T4V
 
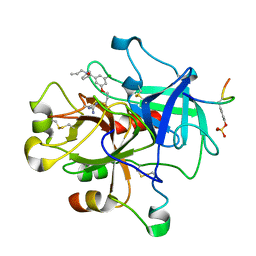 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | Hirudin IIIA, N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
5UFO
 
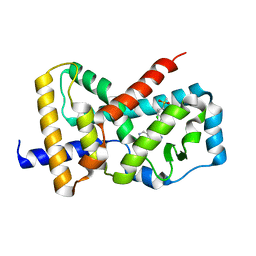 | | Structure of RORgt bound to | | Descriptor: | (S)-{4-chloro-2-methoxy-3-[4-(methylsulfonyl)phenyl]quinolin-6-yl}(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UHI
 
 | | Structure of RORgt bound to | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(4-chlorophenyl)(1-methyl-1H-imidazol-5-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1TJZ
 
 | |
1T4U
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | 2-METHANESULFONYL-BENZENESULFONIC ACID 3-METHYL-5-((1-AMIDINOAMINOOXYMETHYL-CYCLOPROPYL)METHYLOXY)-PHENYLESTER, Hirudin IIIA, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
5UFR
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
5W4R
 
 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | 1-{4-[(R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]piperidin-1-yl}ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5W4V
 
 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Hars, U. | | Deposit date: | 2017-06-13 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1WBK
 
 | |
1IKW
 
 | | Wild Type HIV-1 Reverse Transcriptase in Complex with Efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKX
 
 | |
1IKV
 
 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKY
 
 | | HIV-1 Reverse Transcriptase in Complex with the Inhibitor MSC194 | | Descriptor: | 1-[2-(3-ACETYL-2-HYDROXY-6-METHOXY-PHENYL)-CYCLOPROPYL]-3-(5-CYANO-PYRIDIN-2-YL)-THIOUREA, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
4K5A
 
 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Bcl-2-like protein 2, Designed Ankyrin Repeat Protein 013_D12 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
1W8U
 
 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W6I
 
 | | plasmepsin II-pepstatin A complex | | Descriptor: | PEPSTATIN, PLASMEPSIN 2 PRECURSOR | | Authors: | Lindberg, J, Johansson, P.-O, Rosenquist, A, Kvarnstroem, I, Vrang, L, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-18 | | Release date: | 2006-07-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Study of a Novel Inhibitor with Bulky P1 Side Chain in Complex with Plasmepsin II -Implications for Drug Design
To be Published
|
|
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
