5G37
 
 | | MR structure of the binary mosquito larvicide BinAB at pH 5 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2016-04-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5DXT
 
 | | p110alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, 1,2-ETHANEDIOL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
6VYG
 
 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
3L64
 
 | | T4 Lysozyme S44E/WT* | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | Blaber, M, Zhang, X.-J, Lindstrom, J.D, Pepiot, S.D, Baase, W.A, Matthews, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme.
J.Mol.Biol., 235, 1994
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
3KJQ
 
 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
2YG0
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2YFZ
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
2Y7X
 
 | | The discovery of potent and long-acting oral factor Xa inhibitors with tetrahydroisoquinoline and benzazepine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-(5-FLUORO-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL)-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Adams, C, Belton, D, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Harling, J.D, Irving, W.R, Irvine, S, Kleanthous, S, McLay, I.M, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Discovery of Potent and Long-Acting Oral Factor Xa Inhibitors with Tetrahydroisoquinoline and Benzazepine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2YB7
 
 | | CBM62 in complex with 6-alpha-D-Galactosyl-mannotriose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, McKee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-03-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
1WBP
 
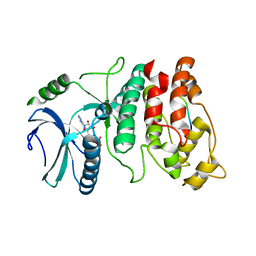 | | SRPK1 bound to 9mer docking motif peptide | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, MEMBRANE-ASSOCIATED GUANYLATE KINASE, ... | | Authors: | Ngo, J.C, Gullinsgrud, J, Chakrabarti, S, Nolen, B, Aubol, B.E, Fu, X.-D, Adams, J.A, McCammon, J.A, Ghosh, G. | | Deposit date: | 2004-11-04 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interplay between Srpk and Clk/Sty Kinases in Phosphorylation of the Splicing Factor Asf/Sf2 is Regulated by a Docking Motif in Asf/Sf2
Mol.Cell, 20, 2005
|
|
5FAL
 
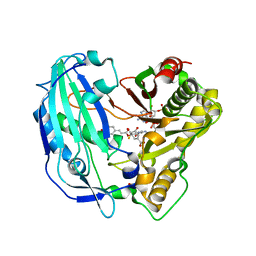 | | Crystal structure of PvHCT in complex with CoA and p-coumaroyl-shikimate | | Descriptor: | (3~{R},4~{R},5~{R})-5-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-3,4-bis(oxidanyl)cyclohexene-1-carboxylic acid, COENZYME A, GLYCEROL, ... | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
5UE3
 
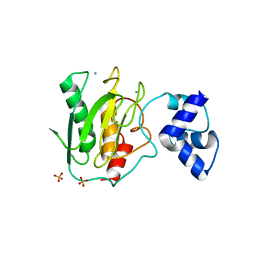 | | proMMP-9desFnII | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Matrix metalloproteinase-9, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
1RTH
 
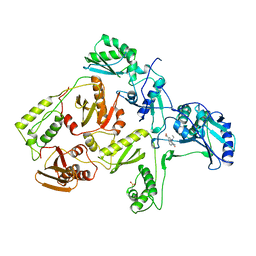 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
2Y6J
 
 | | X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
1U8G
 
 | | Crystal structure of a HIV-1 Protease in complex with peptidomimetic inhibitor KI2-PHE-GLU-GLU-NH2 | | Descriptor: | PROTEASE RETROPEPSIN, peptidomimetic inhibitor KI2-PHE-GLU-GLU-NH2 | | Authors: | Brynda, J, Rezacova, P, Fabry, M, Horejsi, M, Hradilek, M, Soucek, R, Stouracova, R, Konvalinka, J, Sedlacek, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-11-02 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Inhibitor binding at the protein interface in crystals of a HIV-1 protease complex.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7TQY
 
 | | CaKip3[2-482] - ADP-AlFx in complex with a microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Hunter, B, Allingham, J, Sosa, H. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape.
Nat Commun, 13, 2022
|
|
1RL8
 
 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
5FGX
 
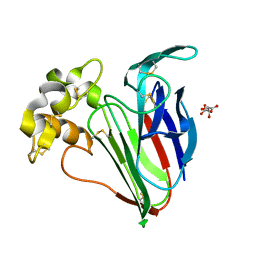 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
5FAN
 
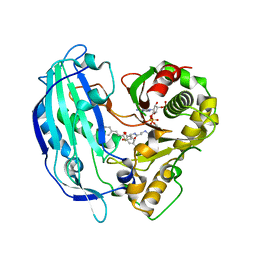 | | Crystal structure of PvHCT in complex with p-coumaroyl-CoA and protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase 2, p-coumaroyl-CoA | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
4B1Q
 
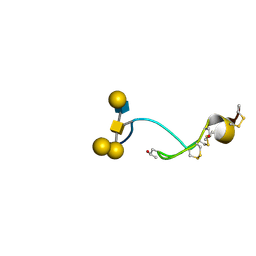 | | NMR structure of the glycosylated conotoxin CcTx from Conus consors | | Descriptor: | CONOTOXIN CCTX, alpha-L-galactopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Hocking, H.G, Gerwig, G.J, Favreau, P, Stocklin, R, Kamerling, J.P, Boelens, R. | | Deposit date: | 2012-07-12 | | Release date: | 2013-02-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the O-Glycosylated Conopeptide Cctx from Conus Consors Venom.
Chemistry, 19, 2013
|
|
5UE4
 
 | | proMMP-9desFnII complexed to JNJ0966 INHIBITOR | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, SULFATE ION, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
6W8S
 
 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
3BMY
 
 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-chloro-6-{5-[(2-morpholin-4-ylethyl)amino]-1,2-benzisoxazol-3-yl}benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
