4D2W
 
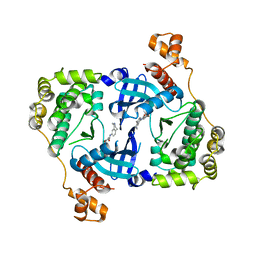 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-bromo-N-(2,3,4,5-tetrahydro-1H-3-benzazepin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-13 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
8VWU
 
 | |
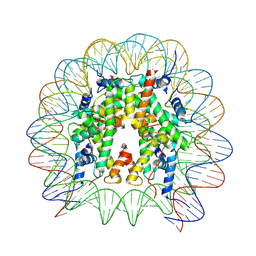
8VWS
 
 | |
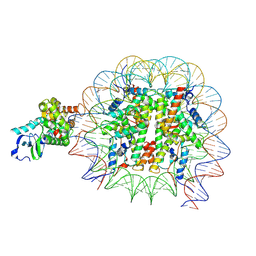
8VWT
 
 | |
8VWV
 
 | |
4A9V
 
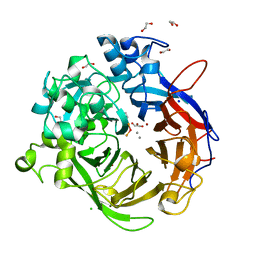 | | Pseudomonas fluorescens PhoX | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-28 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
3PMX
 
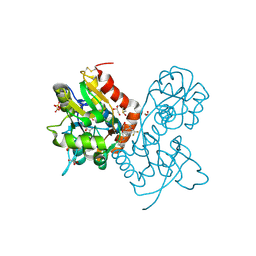 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2FS2
 
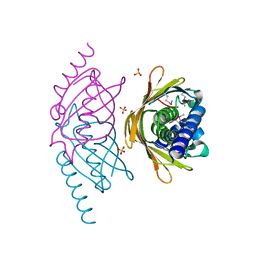 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein paaI, SULFATE ION | | Authors: | Kniewel, R, Buglino, J.A, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, Function, and Mechanism of the Phenylacetate Pathway Hot Dog-fold Thioesterase PaaI
J.Biol.Chem., 281, 2006
|
|
4A9X
 
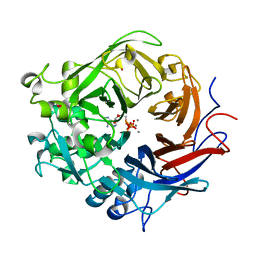 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MU-OXO-DIIRON, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4H3B
 
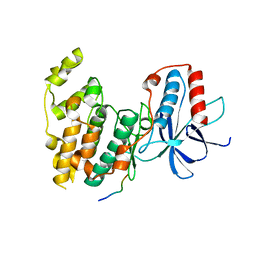 | | Crystal Structure of JNK3 in Complex with SAB Peptide | | Descriptor: | Mitogen-activated protein kinase 10, SH3 domain-binding protein 5 | | Authors: | Nwachukwu, J.C, Laughlin, J.D, Figuera-Losada, M, Cherry, L, Nettles, K.W, LoGrasso, P.V. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Mechanisms of Allostery and Autoinhibition in JNK Family Kinases.
Structure, 20, 2012
|
|
4B1C
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-cyclopropyl-5-methyl-2-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-2H-imidazol-4-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
4B1E
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-methyl-5-phenyl-2-(3-pyridin-3-ylphenyl)-2,3-dihydro-1H-imidazol-4-amine, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
2GC8
 
 | | Structure of a Proline Sulfonamide Inhibitor Bound to HCV NS5b Polymerase | | Descriptor: | 1-[(2-AMINO-4-CHLORO-5-METHYLPHENYL)SULFONYL]-L-PROLINE, RNA-directed RNA polymerase | | Authors: | Gopalsamy, A, Chopra, R, Lim, K, Ciszewski, G, Shi, M, Curran, K.J, Sukits, S.F, Svenson, K, Bard, J, Ellingboe, J.W, Agarwal, A, Krishnamurthy, G, Howe, A.Y, Orlowski, M, Feld, B, O'connell, J, Mansour, T.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Proline Sulfonamides as Potent and Selective Hepatitis C Virus NS5b Polymerase Inhibitors. Evidence for a New NS5b Polymerase Binding Site.
J.Med.Chem., 49, 2006
|
|
4FPR
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, M.H, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2013-12-11 | | Last modified: | 2022-08-03 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
4AMF
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
4ACX
 
 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 23 | | Descriptor: | (8R)-8-[4-(DIFLUOROMETHOXY)PHENYL]-3,3-DIFLUORO-8-[3-(3-METHOXYPROP-1-YN-1-YL)PHENYL]-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4FPQ
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, MH, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
3DWQ
 
 | | Crystal structure of the A-subunit of the AB5 toxin from E. coli with Neu5Gc-2,3Gal-1,3GlcNAc | | Descriptor: | AZIDE ION, N-PROPANOL, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
4GKP
 
 | |
3DWA
 
 | | Crystal structure of the B-subunit of the AB5 toxin from E. coli | | Descriptor: | PENTAETHYLENE GLYCOL, Subtilase cytotoxin, subunit B | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
3DWP
 
 | | Crystal structure of the B-subunit of the AB5 toxin from E. Coli with Neu5Gc | | Descriptor: | N-glycolyl-alpha-neuraminic acid, PENTAETHYLENE GLYCOL, Subtilase cytotoxin, ... | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
3PMW
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | DIMETHYL SULFOXIDE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4BOF
 
 | | Crystal structure of arginine deiminase from group A streptococcus | | Descriptor: | ARGININE DEIMINASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Henningham, A, Ericsson, D.J, Langer, K, Casey, L, Jovcevski, B, Chhatwal, G.S, Aquilina, J.A, Batzloff, M.R, Kobe, B, Walker, M.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus.
MBio, 4, 2013
|
|
3RBU
 
 | | N-terminally AviTEV-tagged Human Glutamate Carboxypeptidase II in complex with 2-PMPA | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tykvart, J, Sacha, P, Barinka, C, Starkova, J, Knedlik, T, Lubkowski, J, Konvalinka, J. | | Deposit date: | 2011-03-30 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Efficient and versatile one-step affinity purification of in vivo biotinylated proteins: Expression, characterization and structure analysis of recombinant human glutamate carboxypeptidase II.
Protein Expr.Purif., 82, 2012
|
|
