6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | 分子名称: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | 著者 | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | 登録日 | 2019-03-07 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | 分子名称: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | 著者 | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | 登録日 | 2019-03-07 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6FCE
 
 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | 分子名称: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | 著者 | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | 登録日 | 2017-12-20 | | 公開日 | 2018-04-25 | | 最終更新日 | 2018-05-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
5ACC
 
 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | 分子名称: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | 著者 | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | 登録日 | 2015-08-15 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | 分子名称: | Urotensin-2 receptor | | 著者 | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | 登録日 | 2018-10-11 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
8G2W
 
 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-06 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G1S
 
 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-02 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4W
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G7E
 
 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-16 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6GJ4
 
 | | Tubulin-6j complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(quinolin-5-yl)naphtho[2,3-b]pyrrolo[1,2-d][1,4]oxazepin-4-yl acetate, CALCIUM ION, ... | | 著者 | Brindisi, M, Ulivieri, C, Alfano, G, Gemma, S, Balaguer, F.d.A, Khan, T, Grillo, A, Chemi, G, Menchon, G, Prota, A.E, Olieric, N, Agell, D.L, Barasoain, I, Diaz, J.F, Nebbioso, A, Conte, M.R, Lopresti, L, Magnano, S, Amet, R, Kinsella, P, Zisterer, D.M, Ibrahim, O, O'Sullivan, J, Morbidelli, L, Spaccapelo, R, Baldari, C, Butini, S, Novellino, E, Campiani, G, Altucci, L, Steinmetz, M.O, Brogi, S. | | 登録日 | 2018-05-16 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-activity relationships, biological evaluation and structural studies of novel pyrrolonaphthoxazepines as antitumor agents.
Eur J Med Chem, 162, 2018
|
|
8G00
 
 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-mer), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-01-31 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6FLM
 
 | |
8PPL
 
 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | 登録日 | 2023-07-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.65 Å) | | 主引用文献 | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
6DZD
 
 | | Crystal structure of Bacillus licheniformis hypothetical protein YfiH | | 分子名称: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | 著者 | Almeida, L.R, Grejo, M.P, Mulinari, E.J, Santos, J.C, Camargo, S, Bernardes, A, Muniz, J.R.C. | | 登録日 | 2018-07-03 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Crystal structure of Bacillus licheniformis hypothetical protein YfiH
To Be Published
|
|
6HUE
 
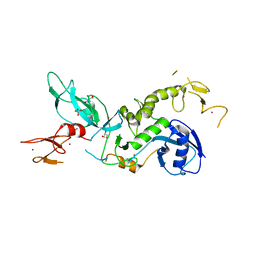 | | ParkinS65N | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | 著者 | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | 登録日 | 2018-10-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
6S2Y
 
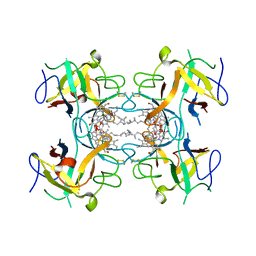 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum with Chlorophyll-b | | 分子名称: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | 著者 | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | 登録日 | 2019-06-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
8PPK
 
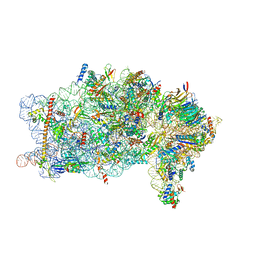 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | 登録日 | 2023-07-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
6S2Z
 
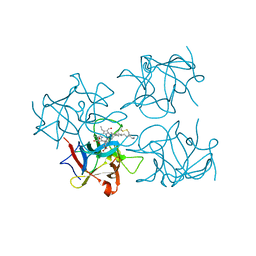 | | Water-soluble Chlorophyll Protein (WSCP) from Brassica oleracea var. Botrytis with Chlorophyll-b | | 分子名称: | CHLOROPHYLL B, Water-Soluble Chlorophyll Protein | | 著者 | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | 登録日 | 2019-06-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
5YVS
 
 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | 分子名称: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | 著者 | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | 登録日 | 2017-11-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.345 Å) | | 主引用文献 | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
6SLZ
 
 | | Crystal structure of human ROR gamma LBD in complex with a (quinolinoxymethyl)benzamide inverse agonist | | 分子名称: | (2~{S})-1-[2,4-bis(chloranyl)-3-[[2-methyl-4-(trifluoromethyl)quinolin-8-yl]oxymethyl]phenyl]carbonyl-~{N}-methyl-pyrrolidine-2-carboxamide, Nuclear receptor ROR-gamma | | 著者 | Amaudrut, J, Argiriadi, M.A, Barth, M, Breinlinger, E.C, Calderwood, D.J, Cusack, K.P, Kort, M.E, Montalbetti, C, Potin, D, Poupardin, O, Spitzer, L. | | 登録日 | 2019-08-21 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of human ROR gamma LBD in complex with a (quinolinoxymethyl)benzamide inverse agonist
To Be Published
|
|
6ERI
 
 | | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10 alpha, ... | | 著者 | Perez Borema, A, Aibara, S, Paul, B, Tobiasson, V, Kimanius, D, Forsberg, B.O, Wallden, K, Lindahl, E, Amunts, A. | | 登録日 | 2017-10-18 | | 公開日 | 2018-04-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor.
Nat Plants, 4, 2018
|
|
6RFE
 
 | | Human protein kinase CK2 alpha in complex with 2-cyano-2-propenamide compound 4 | | 分子名称: | (~{E})-~{N}-(5-bromanyl-1,3,4-thiadiazol-2-yl)-2-cyano-3-(3-methoxy-4-oxidanyl-phenyl)prop-2-enamide, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | 著者 | Dalle Vedove, A, Zanforlin, E, Ribaudo, G, Zagotto, G, Battistutta, R, Lolli, G. | | 登録日 | 2019-04-13 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | A novel class of selective CK2 inhibitors targeting its open hinge conformation.
Eur.J.Med.Chem., 195, 2020
|
|
8QCB
 
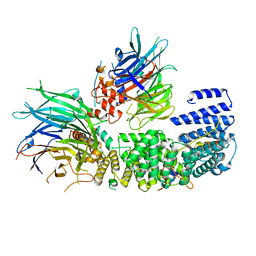 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the open state | | 分子名称: | Antiviral helicase SKI2, Antiviral protein SKI8, Superkiller protein 3, ... | | 著者 | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | 登録日 | 2023-08-25 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8QCA
 
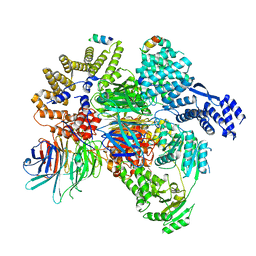 | | CryoEM structure of a S. Cerevisiae Ski2387 complex in the closed state bound to RNA | | 分子名称: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | 著者 | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | 登録日 | 2023-08-25 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
8Q9T
 
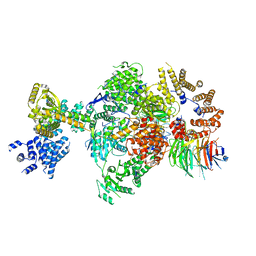 | | CryoEM structure of a S. Cerevisiae Ski238 complex bound to RNA | | 分子名称: | Antiviral helicase SKI2, Antiviral protein SKI8, RNA (5'-R(P*UP*UP*UP*U)-3'), ... | | 著者 | Keidel, A, Koegel, A, Reichelt, P, Kowalinski, E, Schaefer, I.B, Conti, E. | | 登録日 | 2023-08-21 | | 公開日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Mol.Cell, 83, 2023
|
|
