6KDL
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (I) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.274 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
3T8S
 
 | | Apo and InsP3-bound Crystal Structures of the Ligand-Binding Domain of an InsP3 Receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Lin, C, Baek, K, Lu, Z. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Apo and InsP(3)-bound crystal structures of the ligand-binding domain of an InsP(3) receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6KDP
 
 | | Crystal structure of human DNMT3B-DNMT3L complex (II) | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, FORMIC ACID, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDB
 
 | | Crystal structure of human DNMT3B-DNMT3L in complex with DNA containing CpGpT site | | Descriptor: | DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.862 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDT
 
 | | Crystal structure of human DNMT3B (Q772R)-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, FORMIC ACID, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
6KDA
 
 | | Crystal structure of human DNMT3B-DNMT3L in complex with DNA containing CpGpG site | | Descriptor: | DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Lin, C.-C, Chen, Y.-P, Yang, W.-Z, Shen, C.-K, Yuan, H.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Nucleic Acids Res., 48, 2020
|
|
8WVW
 
 | | Cryo-EM structure of LGR4 in state II | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
8WVV
 
 | | Cryo-EM structure of LGR4 in state I | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
6XJ6
 
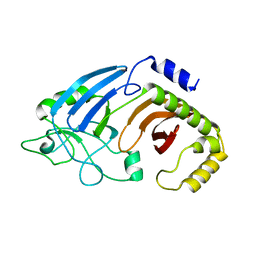 | |
6XJ7
 
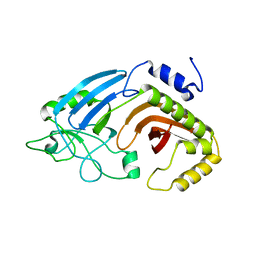 | |
8WVY
 
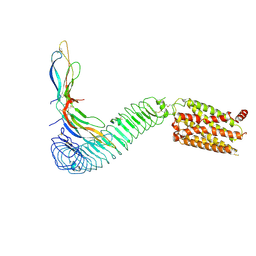 | |
8GLD
 
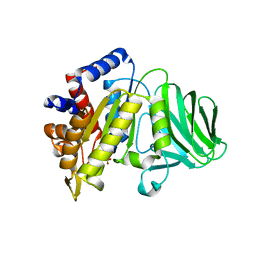 | |
3U1K
 
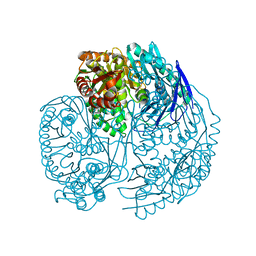 | | Crystal structure of human PNPase | | Descriptor: | CITRIC ACID, Polyribonucleotide nucleotidyltransferase 1, mitochondrial | | Authors: | Lin, C.L, Yuan, H.S. | | Deposit date: | 2011-09-30 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of human polynucleotide phosphorylase: insights into its domain function in RNA binding and degradation
Nucleic Acids Res., 40, 2012
|
|
7EI3
 
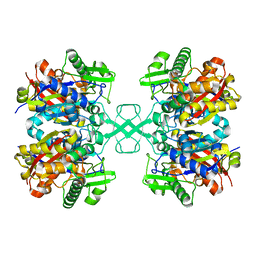 | |
7EI4
 
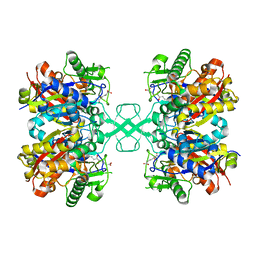 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
1AW4
 
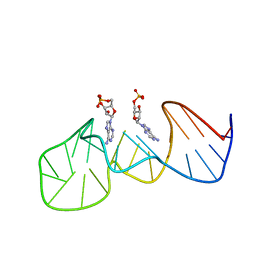 | |
7WQJ
 
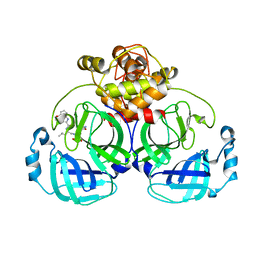 | | Crystal structure of MERS main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7VTC
 
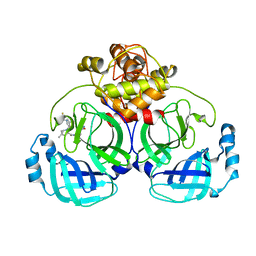 | | Crystal structure of MERS main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53865623 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
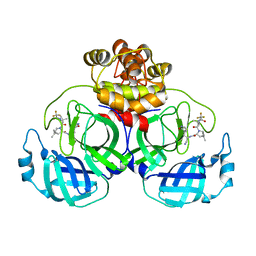 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0227 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7FEA
 
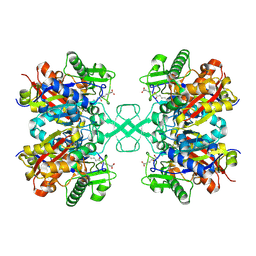 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
7WQI
 
 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
2MG8
 
 | |
4OM5
 
 | | Crystal structure of CTX A4 from Taiwan Cobra (Naja naja atra) | | Descriptor: | Cytotoxin 4 | | Authors: | Lin, C.C, Chang, C.I, Wu, W.G. | | Deposit date: | 2014-01-26 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Endocytotic Routes of Cobra Cardiotoxins Depend on Spatial Distribution of Positively Charged and Hydrophobic Domains to Target Distinct Types of Sulfated Glycoconjugates on Cell Surface.
J.Biol.Chem., 289, 2014
|
|
107D
 
 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | Descriptor: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | Authors: | Lin, C.H, Patel, D.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
4YPM
 
 | |
