1JR7
 
 | |
1KMK
 
 | |
1KMJ
 
 | |
1KPA
 
 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPB
 
 | | PKCI-1-APO | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4M52
 
 | | Structure of Mtb Lpd bound to SL827 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-[(2-amino-5-bromopyridin-3-yl)sulfonyl]-N-(4-methoxyphenyl)-N~2~-methylglycinamide | | Authors: | Lima, C.D. | | Deposit date: | 2013-08-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase.
Biochemistry, 52, 2013
|
|
2NN6
 
 | | Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 | | Descriptor: | 3'-5' exoribonuclease CSL4 homolog, Exosome complex exonuclease RRP4, Exosome complex exonuclease RRP40, ... | | Authors: | Lima, C.D. | | Deposit date: | 2006-10-23 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Reconstitution, activities, and structure of the eukaryotic RNA exosome.
Cell(Cambridge,Mass.), 127, 2006
|
|
2Q2T
 
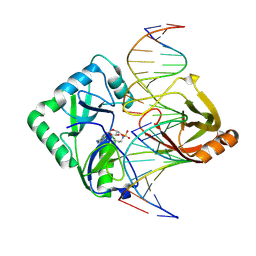 | | Structure of Chlorella virus DNA ligase-adenylate bound to a 5' phosphorylated nick | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', 5'-D(P*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', ... | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1D8H
 
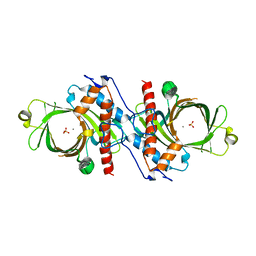 | | X-RAY CRYSTAL STRUCTURE OF YEAST RNA TRIPHOSPHATASE IN COMPLEX WITH SULFATE AND MANGANESE IONS. | | Descriptor: | MANGANESE (II) ION, SULFATE ION, mRNA TRIPHOSPHATASE CET1 | | Authors: | Lima, C.D, Wang, L.K, Shuman, S. | | Deposit date: | 1999-10-24 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus.
Cell(Cambridge,Mass.), 99, 1999
|
|
1D8I
 
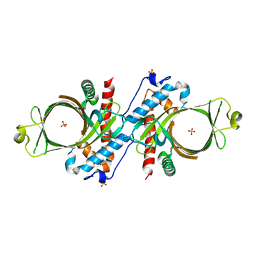 | |
1FIT
 
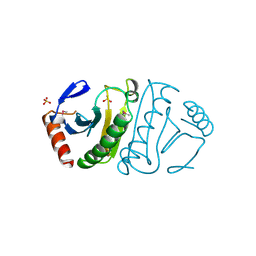 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
4FIT
 
 | | FHIT-APO | | Descriptor: | FRAGILE HISTIDINE TRIAD PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5VZJ
 
 | |
1RI6
 
 | | Structure of a putative isomerase from E. coli | | Descriptor: | putative isomerase ybhE | | Authors: | Lima, C.D, Kniewel, R, Solorzano, V, Wu, J, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative 7-bladed propeller isomerase
To be Published
|
|
2FIT
 
 | | FHIT (FRAGILE HISTIDINE TRIAD PROTEIN) | | Descriptor: | FRAGILE HISTIDINE PROTEIN, SULFATE ION, beta-D-fructofuranose | | Authors: | Lima, C.D, D'Amico, K.L, Naday, I, Rosenbaum, G, Westbrook, E.M, Hendrickson, W.A. | | Deposit date: | 1997-05-17 | | Release date: | 1997-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family.
Structure, 5, 1997
|
|
2HV9
 
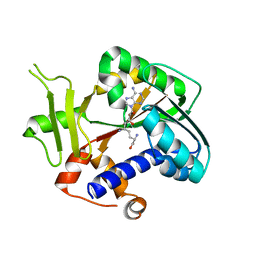 | | Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Lima, C.D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Analysis of Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase, Structure of the Enzyme Bound to Sinefungin, and Evidence That Cap Methyltransferase Is the Target of Sinefungin's Antifungal Activity
J.Biol.Chem., 281, 2006
|
|
2A8X
 
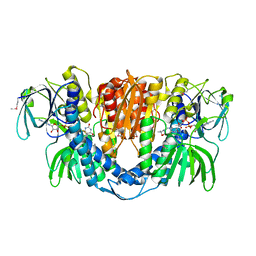 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
2AP9
 
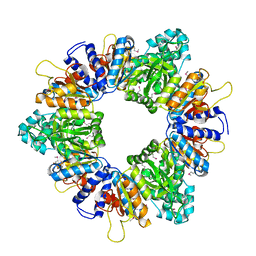 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
6OP8
 
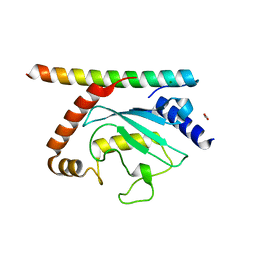 | | S. pombe Ubc7/U7BR complex | | Descriptor: | 1,2-ETHANEDIOL, CUE domain-containing protein 4, mitochondrial, ... | | Authors: | Hann, Z.S, Lima, C.D. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Crystal structure of the Schizosaccharomyces pombe U7BR E2-binding region in complex with Ubc7.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6O83
 
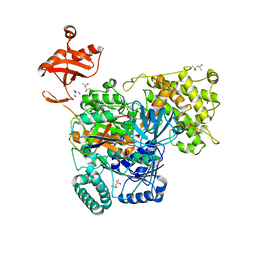 | | S. pombe ubiquitin E1~ubiquitin-AMP tetrahedral intermediate mimic | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, ... | | Authors: | Hann, Z.S, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5FG1
 
 | |
4XQ0
 
 | |
4XPZ
 
 | |
5FG0
 
 | |
