1IC0
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, Nitrosocyanin | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
3NN8
 
 | |
5TTZ
 
 | |
1IBY
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, HEXANE-1,6-DIOL, NITROSOCYANIN | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
1IBZ
 
 | | RED COPPER PROTEIN NITROSOCYANIN FROM NITROSOMONAS EUROPAEA | | Descriptor: | COPPER (II) ION, NITROSOCYANIN | | Authors: | Lieberman, R.L, Arciero, D.M, Hooper, A.B, Rosenzweig, A.C. | | Deposit date: | 2001-03-29 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a novel red copper protein from Nitrosomonas europaea.
Biochemistry, 40, 2001
|
|
5IN9
 
 | | Crystal structure of Grp94 bound to methyl 3-chloro-2-(2-(1-((5-chlorofuran-2-yl)methyl)-1H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, an inhibitor based on the BnIm and Radamide scaffolds. | | Descriptor: | Endoplasmin, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lieberman, R.L, Huard, D.J.E, Kizziah, J.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of Glucose Regulated Protein 94-Selective Inhibitors Based on the BnIm and Radamide Scaffold.
J.Med.Chem., 59, 2016
|
|
6VIO
 
 | |
1YEW
 
 | |
6AOM
 
 | | Structure of molecular chaperone Grp94 bound to selective inhibitor methyl 2-[2-(2-benzylphenyl)ethyl]-3-chloro-4,6-dihydroxybenzoate | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Second Generation Grp94-Selective Inhibitors Provide Opportunities for the Inhibition of Metastatic Cancer.
Chemistry, 23, 2017
|
|
6AOL
 
 | | Structure of molecular chaperone Grp94 bound to selective inhibitor methyl 3-chloro-2-(2-{2-[(4-fluorophenyl)methyl]phenyl}ethyl)-4,6-dihydroxybenzoate | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, methyl 3-chloro-2-(2-{2-[(4-fluorophenyl)methyl]phenyl}ethyl)-4,6-dihydroxybenzoate | | Authors: | Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.764 Å) | | Cite: | Second Generation Grp94-Selective Inhibitors Provide Opportunities for the Inhibition of Metastatic Cancer.
Chemistry, 23, 2017
|
|
2NSX
 
 | | Structure of acid-beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCEROL, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
2NT1
 
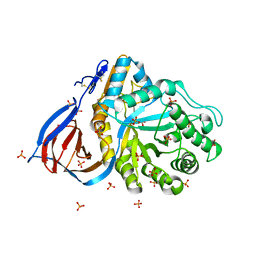 | | Structure of acid-beta-glucosidase at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
2NT0
 
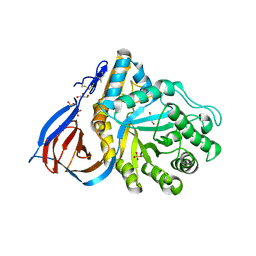 | | Acid-beta-glucosidase low pH, glycerol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glucosylceramidase, ... | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
6PKE
 
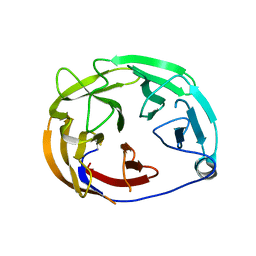 | | Myocilin OLF mutant N428E/D478S | | Descriptor: | Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PKF
 
 | | Myocilin OLF mutant N428E/D478K | | Descriptor: | GLYCEROL, Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PKD
 
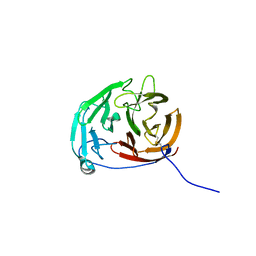 | | Myocilin OLF mutant N428D/D478H | | Descriptor: | Myocilin, SODIUM ION | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5VR2
 
 | |
7SOG
 
 | | Thermostable actophorin | | Descriptor: | Actophorin, NICKEL (II) ION | | Authors: | Lieberman, R.L, Quirk, S. | | Deposit date: | 2021-10-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and activity of a thermally stable mutant of Acanthamoeba actophorin.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6NIZ
 
 | |
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXT
 
 | |
3GXF
 
 | |
3GXI
 
 | | Crystal structure of acid-beta-glucosidase at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXN
 
 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXM
 
 | |
