5GWN
 
 | | Crystal structure of human RCC2 | | Descriptor: | Protein RCC2, SULFATE ION | | Authors: | Liang, L, Yun, C.H, Yin, Y.X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | RCC2 is a novel p53 target in suppressing metastasis.
Oncogene, 37, 2018
|
|
4RZK
 
 | | Crystal structure of Sulfolobus solfataricus Hsp20.1 ACD | | Descriptor: | Small heat shock protein hsp20 family | | Authors: | Liang, L, Yun, C.H. | | Deposit date: | 2014-12-22 | | Release date: | 2015-12-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Crystal structure and function of an unusual dimeric Hsp20.1 provide insight into the thermal protection mechanism of small heat shock proteins.
Biochem.Biophys.Res.Commun., 2015
|
|
4R94
 
 | |
5XDP
 
 | |
3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3F7M
 
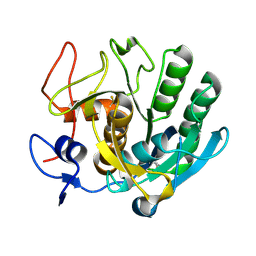 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | Descriptor: | Alkaline serine protease ver112 | | Authors: | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-09 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
6JOJ
 
 | |
6KTO
 
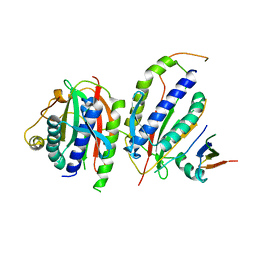 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
6JOI
 
 | |
6JOK
 
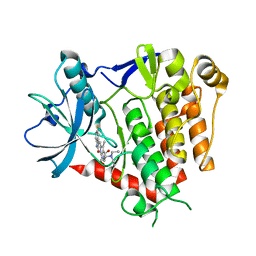 | | Crystal structure of PDGFRA in complex with sunitinib by soaking | | Descriptor: | CHLORIDE ION, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, Platelet-derived growth factor receptor alpha | | Authors: | Liang, L, Yan, X.E, Yun, C.H. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of PDGFRA in complex with sunitinib by soaking
To Be Published
|
|
6JOL
 
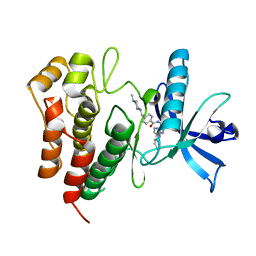 | |
7WM2
 
 | | Cryo-EM structure of AKT1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Dongliang, L, Zijie, Z, Yannan, Q, Yuyue, T, Huaizong, S. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis for the regulation mechanism of a hyperpolarization-activated K+ channel AKT1 by AtKC1
To Be Published
|
|
7WM1
 
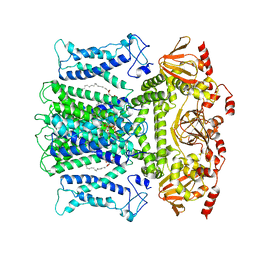 | | Cryo-EM structure of AKT1-AtKC1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium channel AKT1, ... | | Authors: | Dongliang, L, Zijie, Z, Yannan, Q, Yuyue, T, Huaizong, S. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the regulation mechanism of a hyper polarization-activated K+ channel AKT1 by AtKC1
To Be Published
|
|
7MEY
 
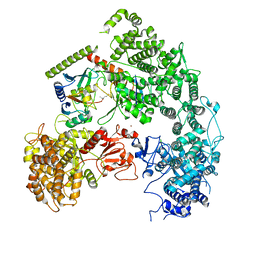 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEX
 
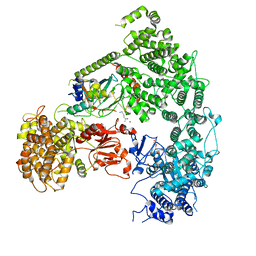 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
2MI7
 
 | | Solution NMR structure of alpha3Y | | Descriptor: | de novo protein a3Y | | Authors: | Glover, S.D, Jorge, C, Liang, L, Valentine, K.G, Hammarstrom, L, Tommos, C. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photochemical tyrosine oxidation in the structurally well-defined alpha 3Y protein: proton-coupled electron transfer and a long-lived tyrosine radical.
J.Am.Chem.Soc., 136, 2014
|
|
5VGT
 
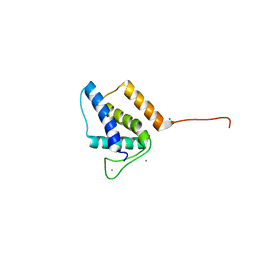 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5K5X
 
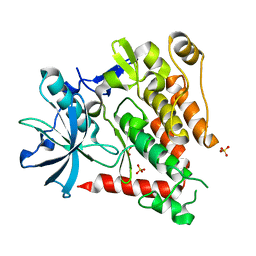 | | Crystal structure of human PDGFRA | | Descriptor: | Platelet-derived growth factor receptor alpha, SULFATE ION | | Authors: | Yan, X.E, Liang, L, Yun, C.H. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural and biochemical studies of the PDGFRA kinase domain
Biochem.Biophys.Res.Commun., 477, 2016
|
|
2LXY
 
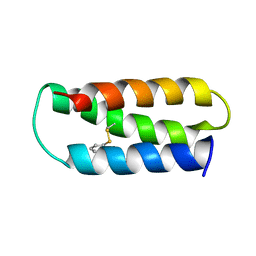 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
9L09
 
 | | SARS-CoV-2 C-RTC with 13-TP | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liang, L, Liu, Y.X, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a 2'-a-Fluoro-2'-b-C-(fluoromethyl) Purine Nucleotide Prodrug as a Potential Oral Anti-SARS-CoV-2 Agent
J Med Chem, 68, 2025
|
|
9INE
 
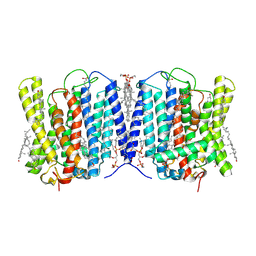 | | Cryo-EM structure of human XPR1 in closed state in the presence of KIDINS220-1-432 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ARACHIDONIC ACID, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9ITG
 
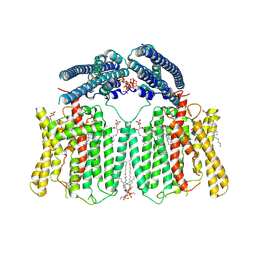 | | Cryo-EM structure of human XPR1-E622A/F623A mutant in complex with InsP6 in inward-facing state in the presence of 10 mM KH2PO4 | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9INF
 
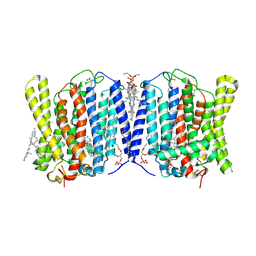 | | Cryo-EM structure of human XPR1 in closed state in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Yin, Y, Zuo, P, Liang, L. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9INH
 
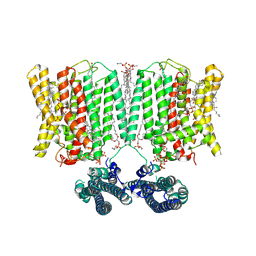 | | Cryo-EM structure of human XPR1 in complex with InsP6 in outward-facing state (SPX visible)-in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9IUC
 
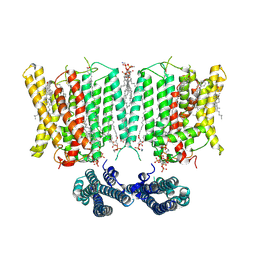 | | Cryo-EM structure of human XPR1 in complex with InsP6 in closed state - in the presence of KIDINS220-1-432 without substrate KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
