1T2T
 
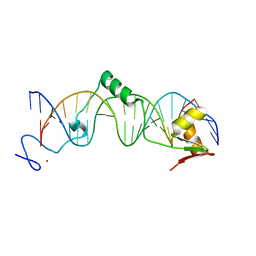 | | Crystal structure of the DNA-binding domain of intron endonuclease I-TevI with operator site | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*AP*GP*TP*CP*CP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*TP*GP*TP*AP*GP*GP*AP*CP*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*T)-3', Intron-associated endonuclease 1, ... | | Authors: | Edgell, D.R, Derbyshire, V, Van Roey, P, LaBonne, S, Stanger, M.J, Li, Z, Boyd, T.M, Shub, D.A, Belfort, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5KGL
 
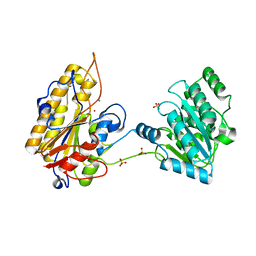 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
1TVI
 
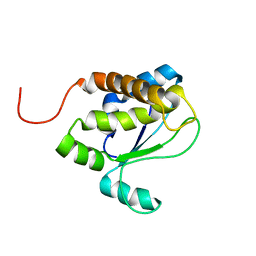 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | Descriptor: | Hypothetical UPF0054 protein TM1509 | | Authors: | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
6Q0V
 
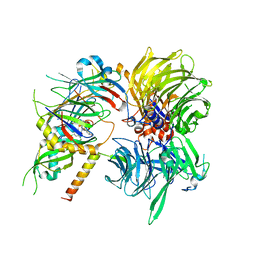 | | Structure of DDB1-DDA1-DCAF15 complex bound to tasisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6OMV
 
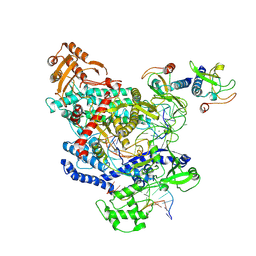 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4-DNA complex | | Descriptor: | AcrVA4, Cpf1, DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*GP*GP*A)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
1FJ8
 
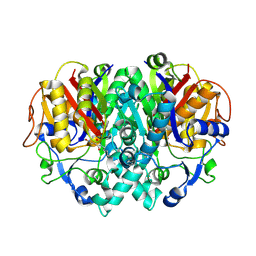 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH CERULENIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1FJ4
 
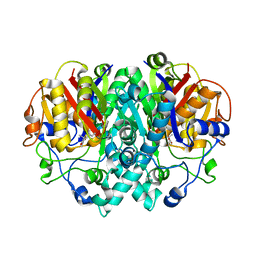 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
4R05
 
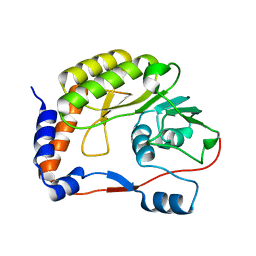 | | Crystal structure of the refolded DENV3 methyltransferase | | Descriptor: | Nonstructural protein NS5 | | Authors: | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
1DBU
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1DBX
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | Descriptor: | cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
7YFU
 
 | | Structure of Rpgrip1l CC2 | | Descriptor: | Protein fantom | | Authors: | He, R, Chen, G, Li, Z, Li, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the N-terminal coiled-coil domains of the ciliary protein Rpgrip1l.
Iscience, 26, 2023
|
|
7YFV
 
 | | Structure of Rpgrip1l CC1 | | Descriptor: | Protein fantom | | Authors: | He, R, Chen, G, Li, Z, Li, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the N-terminal coiled-coil domains of the ciliary protein Rpgrip1l.
Iscience, 26, 2023
|
|
8JJ3
 
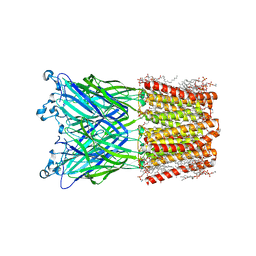 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.6476 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
9ASL
 
 | |
9ASK
 
 | |
9ASJ
 
 | |
8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4
To Be Published
|
|
8ZE6
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to MjDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-04 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of MjHKU4r-CoV-1 RBD bound to MjDPP4
To Be Published
|
|
8ZHE
 
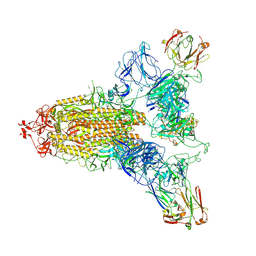 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHP
 
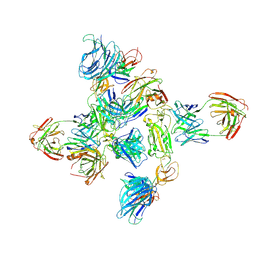 | | Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, Heavy chain of R1-32 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHF
 
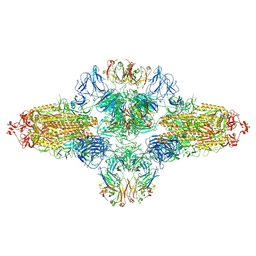 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHH
 
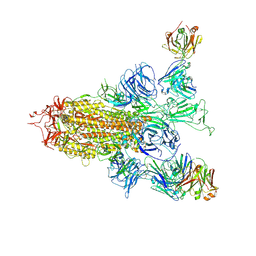 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.55 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHL
 
 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
