3DOE
 
 | | Complex of ARL2 and BART, Crystal Form 1 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
3DOF
 
 | | Complex of ARL2 and BART, Crystal Form 2 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
2C7X
 
 | | Crystal structure of narbomycin-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | CYTOCHROME P450 MONOOXYGENASE, NARBOMYCIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-29 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
7K3V
 
 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7K3W
 
 | | Apoferritin structure at 1.36 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with Falcon4 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.36 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
4DQN
 
 | |
4PED
 
 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
7Y6D
 
 | |
6L8T
 
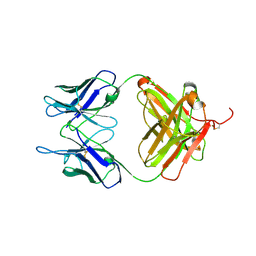 | |
4ORB
 
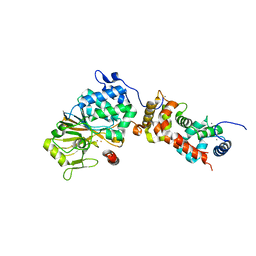 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7XBM
 
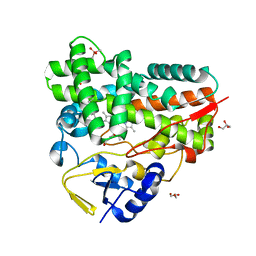 | | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | CACODYLATE ION, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238
Nat Commun, 2023
|
|
5I72
 
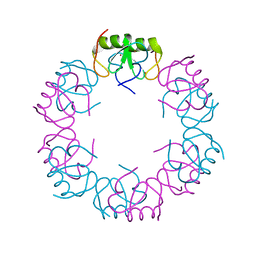 | | Crystal structure of the oligomeric form of the Lassa virus matrix protein Z | | Descriptor: | RING finger protein Z, ZINC ION | | Authors: | Hastie, K, Zandonatti, M, Liu, T, Li, S, Woods Jr, V, Saphire, E.O. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-09 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Oligomeric Form of Lassa Virus Matrix Protein Z.
J.Virol., 90, 2016
|
|
6ONU
 
 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
