4JCL
 
 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
3EXE
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
7C84
 
 | | Esterase AlinE4 mutant, D162A | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
3EXH
 
 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
7YL2
 
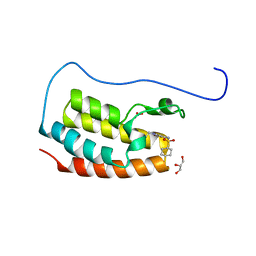 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
3UC4
 
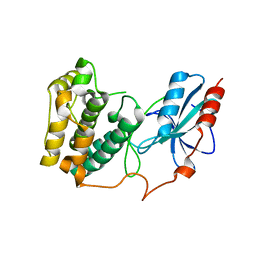 | | The crystal structure of Snf1-related kinase 2.6 | | Descriptor: | Serine/threonine-protein kinase SRK2E | | Authors: | Zhou, X.E, Ng, L.-M, Soon, F.-F, Kovach, A, Suino-Powell, K.M, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-10-26 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3FVX
 
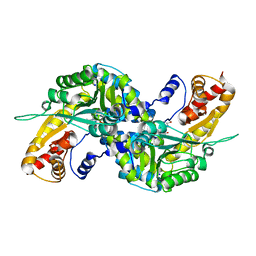 | | Human kynurenine aminotransferase I in complex with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
4QBL
 
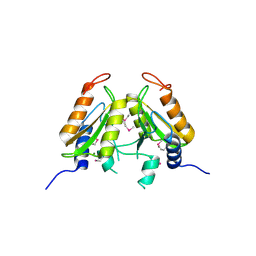 | | VRR_NUC domain protein | | Descriptor: | MAGNESIUM ION, VRR-NUC | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
4LAD
 
 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4QBN
 
 | | VRR_NUC domain | | Descriptor: | Nuclease, SULFATE ION | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
4LJY
 
 | | Crystal structure of RNA splicing effector Prp5 in complex with ADP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
4LK2
 
 | | Crystal structure of RNA splicing effector Prp5 | | Descriptor: | NICKEL (II) ION, Pre-mRNA-processing ATP-dependent RNA helicase PRP5 | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
3KL4
 
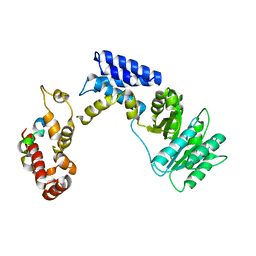 | | Recognition of a signal peptide by the signal recognition particle | | Descriptor: | Signal peptide of yeast dipeptidyl aminopeptidase B, Signal recognition 54 kDa protein | | Authors: | Janda, C.Y, Nagai, K, Li, J, Oubridge, C. | | Deposit date: | 2009-11-06 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Recognition of a signal peptide by the signal recognition particle.
Nature, 465, 2010
|
|
4MLO
 
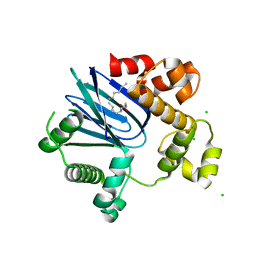 | | 1.65A resolution structure of ToxT from Vibrio cholerae (P21 Form) | | Descriptor: | CHLORIDE ION, PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | Authors: | Lovell, S, Wehmeyer, G, Battaile, K.P, Li, J, Egan, S. | | Deposit date: | 2013-09-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 angstrom resolution structure of the AraC-family transcriptional activator ToxT from Vibrio cholerae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3L3Z
 
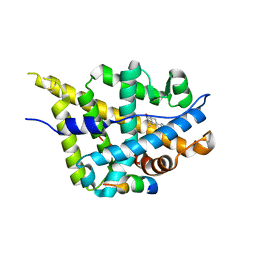 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
5U2U
 
 | |
4E02
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
5SV7
 
 | | The Crystal structure of a chaperone | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4E00
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP
To be Published
|
|
4E01
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with AMPPNP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
3K40
 
 | | Crystal structure of Drosophila 3,4-dihydroxyphenylalanine decarboxylase | | Descriptor: | Aromatic-L-amino-acid decarboxylase, GLYCEROL | | Authors: | Han, Q, Ding, H, Robinson, H, Christensen, B.M, Li, J. | | Deposit date: | 2009-10-05 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
Plos One, 5, 2010
|
|
4FD6
 
 | |
4FD7
 
 | | Crystal structure of insect putative arylalkylamine N-Acetyltransferase 7 from the yellow fever mosquito Aedes aegypt | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, SULFATE ION, ... | | Authors: | Han, Q, Robinson, R, Li, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of insect arylalkylamine N-acetyltransferases: structural evidence from the yellow fever mosquito, Aedes aegypti.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FD5
 
 | |
4FD4
 
 | |
