6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
5YC4
 
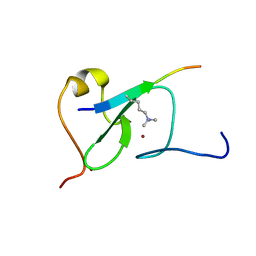 | |
5YC3
 
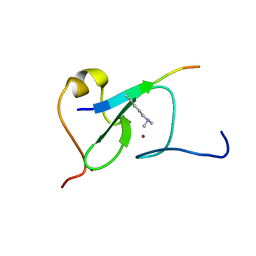 | |
5YKI
 
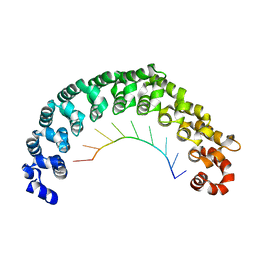 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | Descriptor: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
5YKH
 
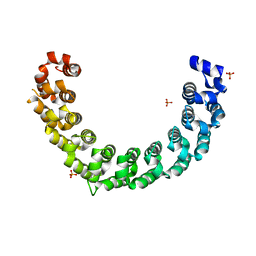 | | Crystal structure of the engineered nine-repeat PUF domain | | Descriptor: | PHOSPHATE ION, Pumilio homolog 1 | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
7WHR
 
 | |
5XFO
 
 | | Structure of the N-terminal domains of PHF1 | | Descriptor: | PHD finger protein 1, ZINC ION | | Authors: | Wang, Z, Li, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polycomb-like proteins link the PRC2 complex to CpG islands
Nature, 549, 2017
|
|
5Z93
 
 | |
6KDS
 
 | |
6L94
 
 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XFN
 
 | | Structure of the N-terminal domains of PHF1 | | Descriptor: | PHD finger protein 1, ZINC ION | | Authors: | Wang, Z, Li, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polycomb-like proteins link the PRC2 complex to CpG islands
Nature, 549, 2017
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
7E9M
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep3 | | Descriptor: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2021-03-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
7EA1
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep2 | | Descriptor: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
4DFU
 
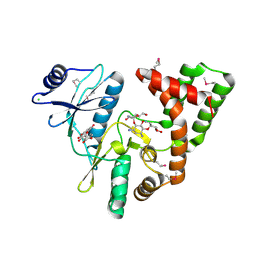 | | Inhibition of an antibiotic resistance enzyme: crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA in complex with kanamycin inhibited with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, APH(2")-Id, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
2RSX
 
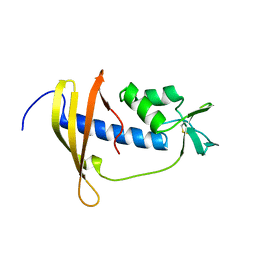 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
8KCO
 
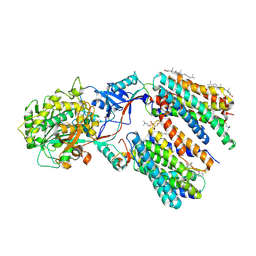 | | Cryo-EM structure of human gamma-secretase in complex with RO4929097 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,2-dimethyl-N-[(7S)-6-oxo-5,7-dihydrobenzo[d][1]benzazepin-7-yl]-N'-(2,2,3,3,3-pentafluoropropyl)propanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Li, H, Kai, U, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560
To Be Published
|
|
4DBX
 
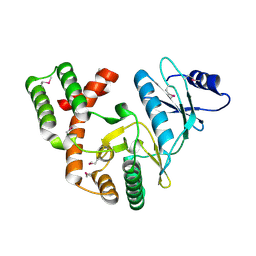 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-ID/APH(2")-IVA | | Descriptor: | APH(2")-ID | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Singer, A.U, Evdokimova, E, Egorova, E, Di Leo, R, Li, H, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-16 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4DFB
 
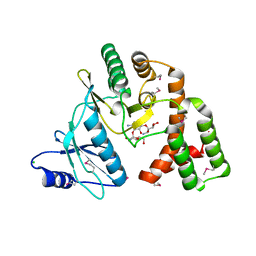 | | Crystal structure of aminoglycoside phosphotransferase aph(2")-id/aph(2")-iva in complex with kanamycin | | Descriptor: | APH(2")-Id, CHLORIDE ION, KANAMYCIN A | | Authors: | Stogios, P.J, Minasov, G, Osipiuk, J, Evdokimova, E, Egorova, E, Di leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
2ECC
 
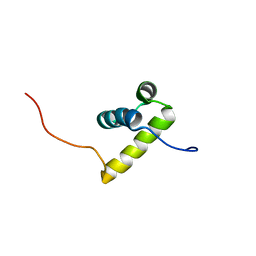 | | Solution Structure of the second Homeobox Domain of Human Homeodomain Leucine Zipper-Encoding Gene (Homez) | | Descriptor: | Homeobox and leucine zipper protein Homez | | Authors: | Ohnishi, S, Kamatari, Y.O, Tochio, N, Nameki, N, Miyamoto, K, Li, H, Kobayashi, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the second Homeobox Domain of Human Homeodomain Leucine Zipper-Encoding Gene (Homez)
To be Published
|
|
3IYM
 
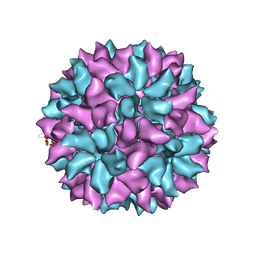 | | Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling | | Descriptor: | Capsid protein | | Authors: | Tang, J, Pan, J, Havens, W.F, Ochoa, W.F, Li, H, Sinkovits, R.S, Guu, T.S.Y, Ghabrial, S.A, Nibert, M.L, Tao, J.Y, Baker, T.S. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Backbone Trace of Partitivirus Capsid Protein from Electron Cryomicroscopy and Homology Modeling
Biophys.J., 99, 2010
|
|
4DE4
 
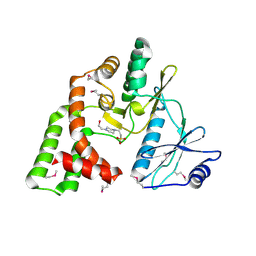 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
5ZMR
 
 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
