7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
4O5A
 
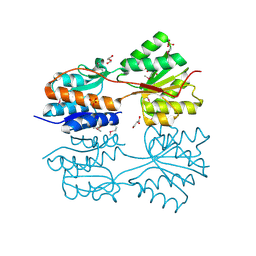 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4L4G
 
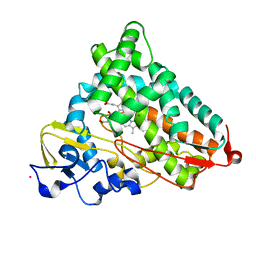 | | Structure of cyanide and camphor bound P450cam mutant L358P/K178G | | Descriptor: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
3UR3
 
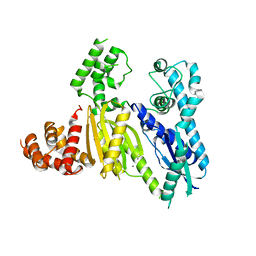 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | CALCIUM ION, Cmr2dHD, ZINC ION | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
3UPS
 
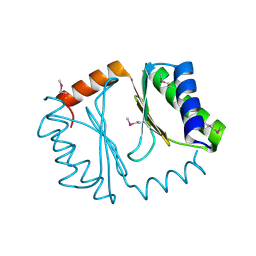 | |
4JS3
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057 | | Descriptor: | 2-chloro-4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4MVE
 
 | |
3UNG
 
 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Cmr2dHD, ... | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
4JTT
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 066 | | Descriptor: | 2,4-dimethyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Zhu, J, Ren, X. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
TO BE PUBLISHED
|
|
4L4C
 
 | | Structure of L358P/K178G mutant of P450cam bound to camphor | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
4MZF
 
 | | Crystal structure of human Spindlin1 bound to histone H3(K4me3-R8me2a) peptide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Peptide from Histone H3.2, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
4JGD
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016 | | Descriptor: | 1-[4-methyl-2-(naphthalen-2-ylamino)-1,3-thiazol-5-yl]ethanone, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A016
TO BE PUBLISHED
|
|
8INR
 
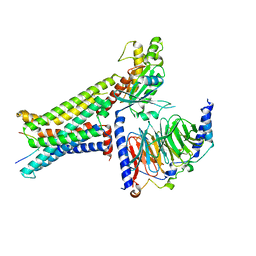 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
3V5S
 
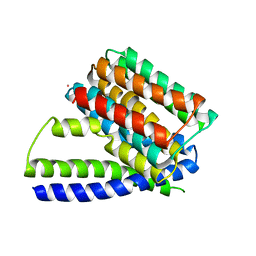 | | Structure of Sodium/Calcium Exchanger from Methanococcus jannaschii | | Descriptor: | CADMIUM ION, Sodium/Calcium Exchanger | | Authors: | Liao, J, Li, H, Zeng, W, Sauer, D.B, Belmares, R, Jiang, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
8IRT
 
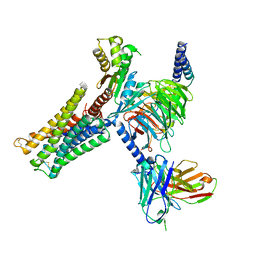 | | Dopamine Receptor D3R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRS
 
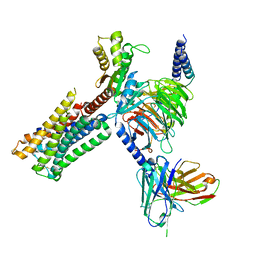 | | Dopamine Receptor D2R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
4JTU
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with brequinar analogue | | Descriptor: | 6-bromo-2-{4-[(2R)-butan-2-yl]phenyl}-3-methylquinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
4JTS
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 072 | | Descriptor: | 4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4LZK
 
 | |
4PE6
 
 | | Crystal structure of ABC transporter solute binding protein from Thermobispora bispora DSM 43833 | | Descriptor: | (2R,3S)-2,3,4-trihydroxybutanoic acid, Putative ABC transporter | | Authors: | Chang, C, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of ABC transporter solute binding protein from Thermobispora bispora DSM 43833
to be published
|
|
4K8F
 
 | |
