8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6U8Y
 
 | |
2AEE
 
 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, GLYCEROL, Orotate phosphoribosyltransferase, ... | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus pyogenes
To be Published
|
|
3JU8
 
 | | Crystal Structure of Succinylglutamic Semialdehyde Dehydrogenase from Pseudomonas aeruginosa. | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of Succinylglutamic Semialdehyde Dehydrogenase from Pseudomonas aeruginosa
To be Published
|
|
4N19
 
 | | Structural basis of conformational transitions in the active site and 80 s loop in the FK506 binding protein FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Mustafi, S.M, Brecher, M.B, Zhang, J, Li, H.M, Lemaster, D.M, Hernandez, G. | | Deposit date: | 2013-10-03 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of conformational transitions in the active site and 80's loop in the FK506-binding protein FKBP12.
Biochem.J., 458, 2014
|
|
7RD6
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD8
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E1-ATP state | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD7
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
4M0M
 
 | | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | PHOSPHATE ION, Putative uncharacterized protein, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Li, H, Clancy, S, Shuman, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | The crystal structure of a functionally unknown protein lpg2422 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
8D4X
 
 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
1ZWZ
 
 | |
7X8G
 
 | |
7X8B
 
 | |
3KBQ
 
 | |
7X88
 
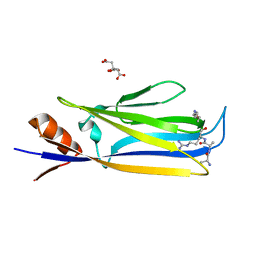 | |
8D2P
 
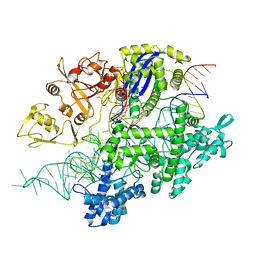 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA target strand (5'-D(P*CP*CP*AP*GP*GP*AP*TP*CP*TP*TP*GP*CP*CP*AP*TP*CP*CP*TP*AP*CP*CP*TP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2K
 
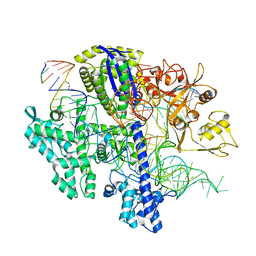 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2O
 
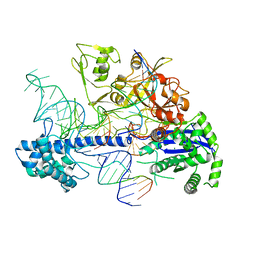 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2L
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2Q
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
5WM9
 
 | |
