7DZK
 
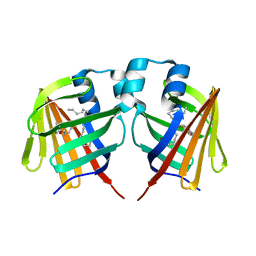 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
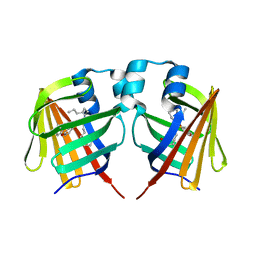 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
6L1R
 
 | |
6L1E
 
 | |
7F7M
 
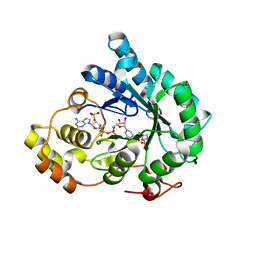 | | AKR4C17 in complex with NADP+ and glyphosate | | Descriptor: | AKR4-2, GLYPHOSATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7L
 
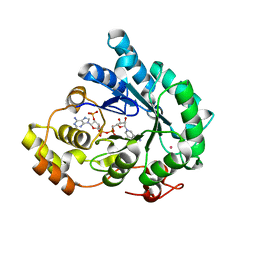 | | Crystal structure of AKR4C17 bound with NADPH | | Descriptor: | AKR4-2, COBALT (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7K
 
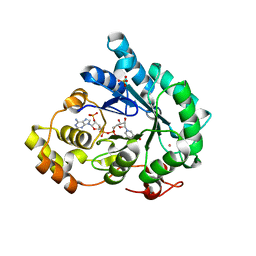 | | Crystal structure of AKR4C17 bound with NADP+ | | Descriptor: | AKR4-2, COBALT (II) ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7DLB
 
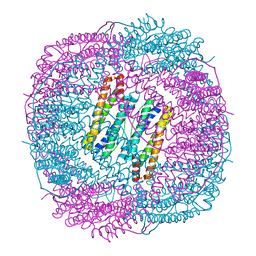 | |
7F7J
 
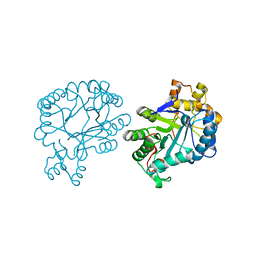 | | The crystal structure of AKR4C17 | | Descriptor: | AKR4-2, COBALT (II) ION, SULFATE ION | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
1R5I
 
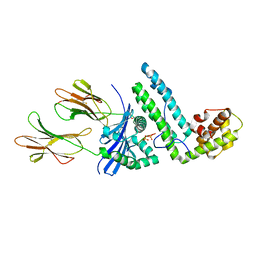 | | Crystal structure of the MAM-MHC complex | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
4N4G
 
 | |
3KPH
 
 | |
4MO7
 
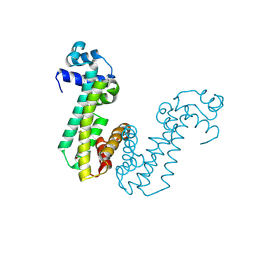 | | Crystal structure of superantigen PfiT | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Transcriptional regulator I2 | | Authors: | Liu, L.H, Chen, H, Li, H.M. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Pfit is a structurally novel Crohn's disease-associated superantigen.
Plos Pathog., 9, 2013
|
|
3B3O
 
 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
1HV8
 
 | |
6ED3
 
 | | Mtb ClpB in complex with AMPPNP | | Descriptor: | Chaperone protein ClpB | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MCA
 
 | |
3JWW
 
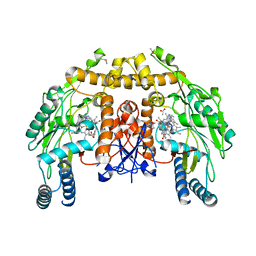 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-[(3'S,4'S)-4'-((6"-amino-4"-methylpyridin-2"-yl)methyl)pyrrolidin-3'-yl]-N2- (3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX5
 
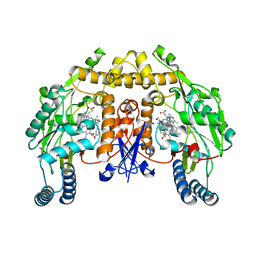 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'S,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
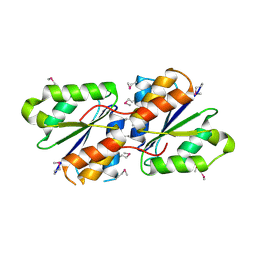
3B48
 
 | |
3JWV
 
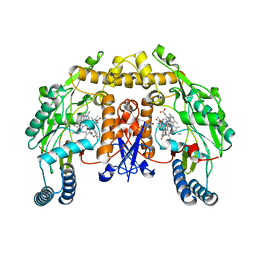 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'S,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
1Q9U
 
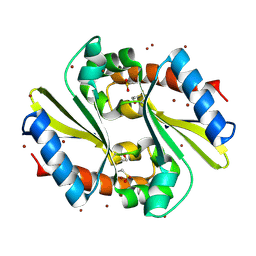 | |
2QMW
 
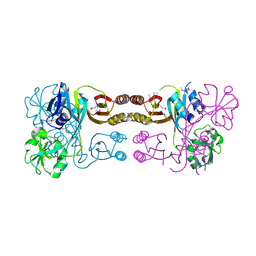 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
