2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
2Q1O
 
 | |
2Q1R
 
 | | Crystal Structure Analysis of the RNA Dodecamer CGCGAAUUAGCG, with a G-A mismatch. | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Li, F, Pallan, P.S. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme.
Nucleic Acids Res., 35, 2007
|
|
5WQ4
 
 | | Crystal structure of OPTN and linear diubiquitin complex | | Descriptor: | Optineurin, ubiquitin | | Authors: | Li, F, Pan, L. | | Deposit date: | 2016-11-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation.
Autophagy, 14, 2018
|
|
6MDW
 
 | | Mechanism of protease dependent DPC repair | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Li, F, Raczynska, J, Chen, Z, Yu, H. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Cell Rep, 26, 2019
|
|
4ZRI
 
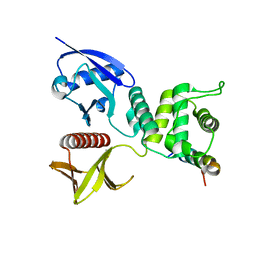 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
6WSL
 
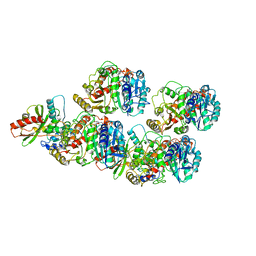 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
7WYG
 
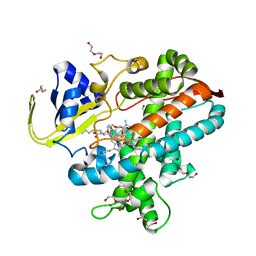 | | Crystal structure of P450BSbeta-L78I/Q85H/G290I variant in complex with palmitic acid. | | Descriptor: | Cytochrome P450 152A1, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Li, F, He, C, Wang, X. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic Enantioselective beta-Hydroxylation of Unactivated C-H Bonds in Aliphatic Carboxylic Acids.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8EON
 
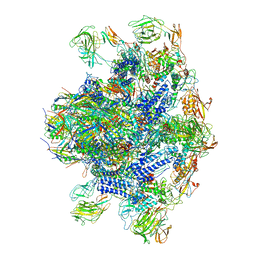 | | Pseudomonas phage E217 baseplate complex | | Descriptor: | Baseplate component gp33, Baseplate component gp34, Baseplate component gp36, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8ENV
 
 | | In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map | | Descriptor: | Baseplate_J domain-containing protein gp44, Ripcord gp36, Sheath initiator gp34, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8FUV
 
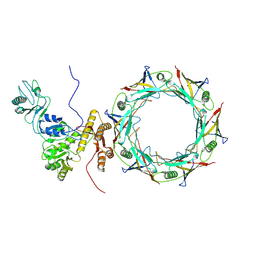 | |
8FVG
 
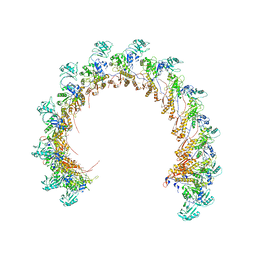 | |
8FRS
 
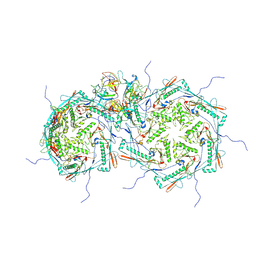 | |
8FVH
 
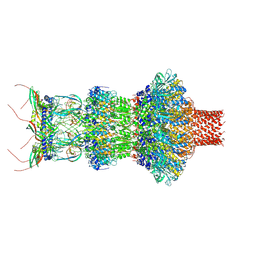 | | Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins) | | Descriptor: | E217 collar protein gp28, E217 gateway protein gp29, E217 head-to-tail connector protein gp27, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZI
 
 | | Crystal Structure of TR3 LBD in complex with DPDO | | Descriptor: | 1-(3,5-dimethoxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4RZF
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4RZG
 
 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
8UCD
 
 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | Authors: | Li, F, Bailis, J.M, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
3UIU
 
 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
4ROU
 
 | |
4UC2
 
 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
4UC1
 
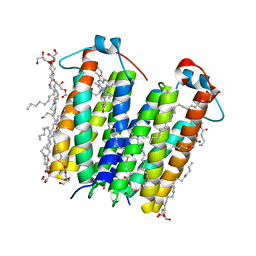 | | High resolution crystal structure of translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, METHOXY-ETHOXYL, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
