5Z2I
 
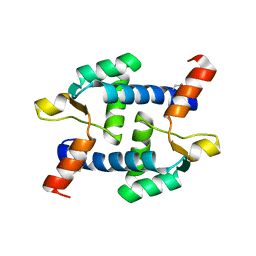 | | Structure of Dictyostelium discoideum mitochondrial calcium uniporter N-ternimal domain (Se-DdMCU-NTD) | | Descriptor: | Dictyostelium discoideum mitochondrial calcium uniporter | | Authors: | Yuan, Y, Wen, M, Chou, J, Li, D, Bo, O. | | Deposit date: | 2018-01-02 | | Release date: | 2019-01-02 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Characterization of the N-Terminal Domain of theDictyostelium discoideumMitochondrial Calcium Uniporter.
Acs Omega, 5, 2020
|
|
2AE9
 
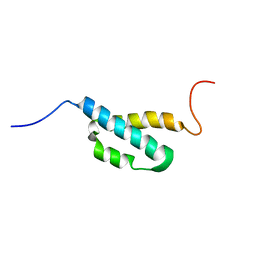 | | Solution Structure of the theta subunit of DNA polymerase III from E. coli | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Mueller, G.A, Kirby, T.W, Derose, E.F, Li, D, Schaaper, R.M, London, R.E. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
J.Bacteriol., 187, 2005
|
|
8I24
 
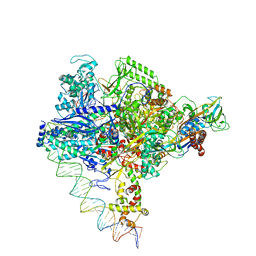 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8IKP
 
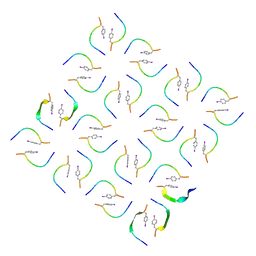 | |
8IK7
 
 | |
8IKB
 
 | |
8IKS
 
 | |
8J03
 
 | |
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
8IY7
 
 | |
8IZZ
 
 | |
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7D2Z
 
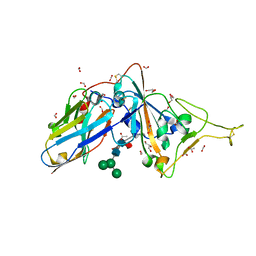 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7D30
 
 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
4NC3
 
 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
8PPO
 
 | |
8Q7P
 
 | |
8Q7M
 
 | |
8Q27
 
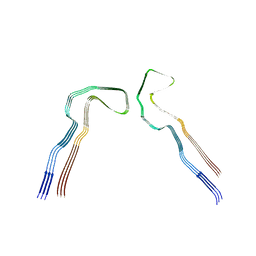 | | Tau: AD-MIA1 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W, Goedert, M, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q2J
 
 | | Tau - AD-MIA2 | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-02 | | Release date: | 2023-08-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q9G
 
 | |
