1FKH
 
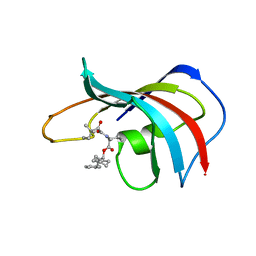 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1FKG
 
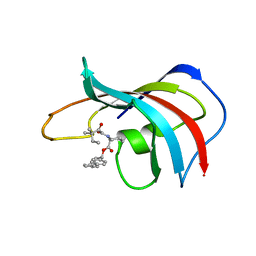 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1,3-DIPHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1FKI
 
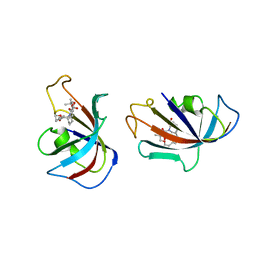 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | (21S)-1AZA-4,4-DIMETHYL-6,19-DIOXA-2,3,7,20-TETRAOXOBICYCLO[19.4.0] PENTACOSANE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
1BGO
 
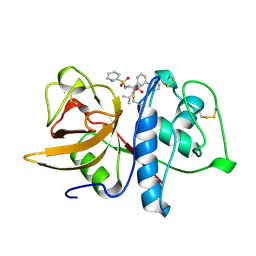 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PEPTIDOMIMETIC INHIBITOR | | Descriptor: | 1-[2-(3-BIPHENYL)-4-METHYLVALERYL)]AMINO-3-(2-PYRIDYLSULFONYL)AMINO-2-PROPANONE, CATHEPSIN K | | Authors: | Desjarlais, R.L, Yamashita, D.S, Oh, H.-J, Bondinell, W.E, Uzinskas, I.N, Erhard, K.F, Allen, A.C, Haltiwanger, R.C, Zhao, B, Smith, W.W, Abdel-Meguid, S.S, D'Alessio, K, Janson, C.A, Mcqueney, M.S, Tomaszek, T.A, Levy, M.A, Veber, D.F. | | Deposit date: | 1998-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of X-Ray Co-Crystal Structures and Molecular Modeling to Design Potent and Selective Non-Peptide Inhibitors of Cathepsin K
J.Am.Chem.Soc., 120, 1998
|
|
3RDX
 
 | | Crystal structure of ligand-free R7-2 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDM
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin/PEG | | Descriptor: | BIOTIN, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDU
 
 | | Crystal structure of R7-2 streptavidin complexed with PEG | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDQ
 
 | | Crystal structure of R7-2 streptavidin complexed with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE6
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | Descriptor: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDS
 
 | | Crystal structure of the refolded R7-2 streptavidin | | Descriptor: | PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3THT
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
