6VZ4
 
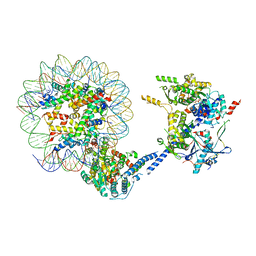 | |
6VZG
 
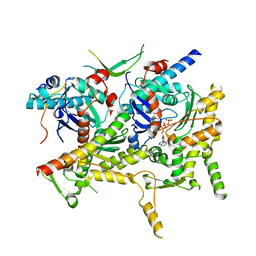 | | Cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Leschziner, A.E, Baker, R.W. | | Deposit date: | 2020-02-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into assembly and function of the RSC chromatin remodeling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9CHO
 
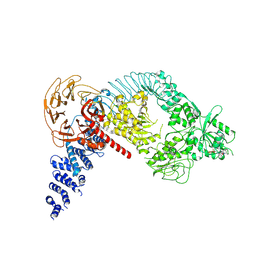 | | Autoinhibited full-length LRRK2(I2020T) on microtubules with MLi-2 | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Chen, S, Villa, E, Leschziner, A.E. | | Deposit date: | 2024-07-01 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron tomography reveals the microtubule-bound form of inactive LRRK2.
Biorxiv, 2024
|
|
6OXL
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 (mu E302K) BOUND TO NECAP IN THE PRESENCE OF SS DNA | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
6OWO
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
7MGM
 
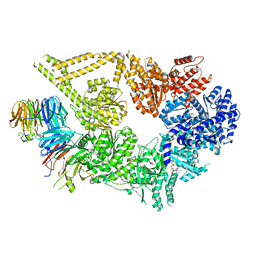 | | Structure of yeast cytoplasmic dynein with AAA3 Walker B mutation bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lahiri, I, Reimer, J.M, Leschziner, A.E. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for cytoplasmic dynein-1 regulation by Lis1.
Elife, 11, 2022
|
|
7THZ
 
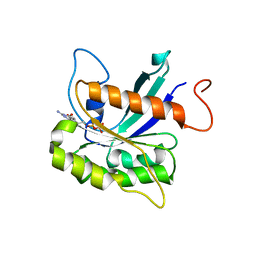 | |
7THY
 
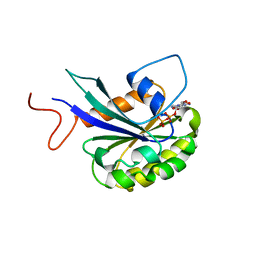 | |
6DRV
 
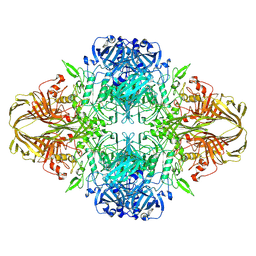 | |
7RS5
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to Taxol-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2021
|
|
3J1U
 
 | | Low affinity dynein microtubule binding domain - tubulin complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | Authors: | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
3J1T
 
 | | High affinity dynein microtubule binding domain - tubulin complex | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | Authors: | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2012-06-25 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
8DYV
 
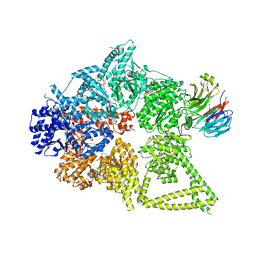 | | Structure of human cytoplasmic dynein-1 bound to one Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Reimer, J.M, DeSantis, M, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2022-08-05 | | Release date: | 2023-02-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structures of human dynein in complex with the lissencephaly 1 protein, LIS1.
Elife, 12, 2023
|
|
8DYU
 
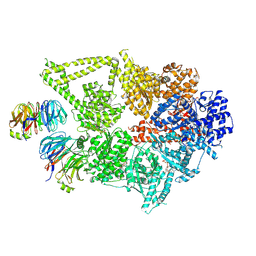 | | Structure of human cytoplasmic dynein-1 bound to two Lis1 proteins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Reimer, J.M, DeSantis, M, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2022-08-05 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human dynein in complex with the lissencephaly 1 protein, LIS1.
Elife, 12, 2023
|
|
8E00
 
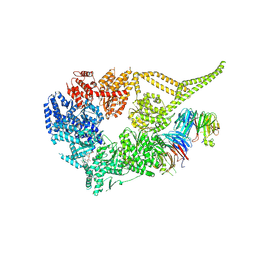 | | Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DZZ
 
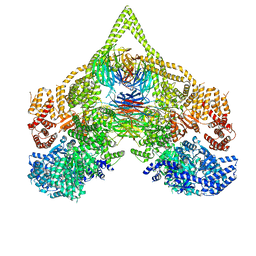 | | Cryo-EM structure of chi dynein bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E06
 
 | | Symmetry expansion of dimeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Lin, Y.X, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E05
 
 | | Structure of dimeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Lin, Y.X, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E04
 
 | | Structure of monomeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Mathea, S, Chatterjee, D, Knapp, S, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6V9H
 
 | | Ankyrin repeat and SOCS-box protein 9 (ASB9), ElonginB (ELOB), and ElonginC (ELOC) bound to its substrate Brain-type Creatine Kinase (CKB) | | Descriptor: | Ankyrin repeat and SOCS box protein 9, Creatine kinase B-type, Elongin-B, ... | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
6V9I
 
 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
7RS6
 
 | | Cryo-EM structure of Kip3 (AMPPNP) bound to GMPCPP-Stabilized Microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hernandez-Lopez, R.A, Leschziner, A.E, Arellano-Santoyo, H, Pellman, D, Stokasimov, E, Wang, R.Y.-R. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Multimodal tubulin binding by the yeast kinesin-8, Kip3, underlies its motility and depolymerization
Biorxiv, 2024
|
|
5VVR
 
 | |
5VVS
 
 | | RNA pol II elongation complex | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leschziner, A.E. | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural basis for the initiation of eukaryotic transcription-coupled DNA repair.
Nature, 551, 2017
|
|
5VLJ
 
 | | Cryo-EM structure of yeast cytoplasmic dynein with Walker B mutation at AAA3 in presence of ATP-VO4 | | Descriptor: | Dynein heavy chain, cytoplasmic, Nuclear distribution protein PAC1 | | Authors: | Cianfrocco, M.A, DeSantis, M.E, Htet, Z.M, Tran, P.T, Reck-Peterson, S.L, Leschziner, A.E. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Lis1 Has Two Opposing Modes of Regulating Cytoplasmic Dynein.
Cell, 170, 2017
|
|
