6NF4
 
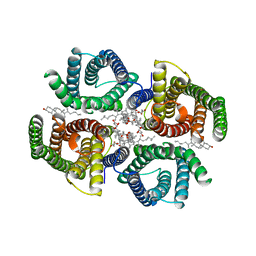 | | Structure of zebrafish Otop1 in nanodiscs | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, Otopetrin1 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NF6
 
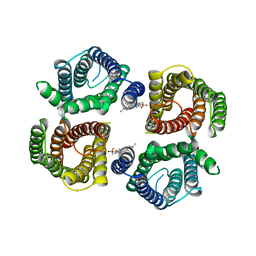 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7CAZ
 
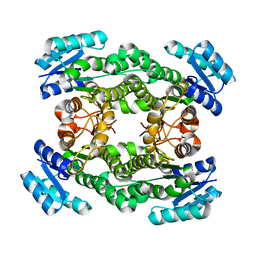 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CFZ
 
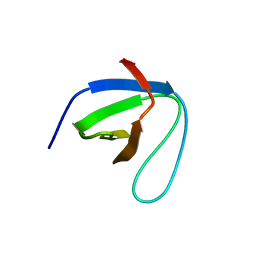 | | SH3 domain of NADPH oxidase activator 1 | | Descriptor: | NADPH oxidase activator 1 | | Authors: | Kim, M, Park, J.H, Attri, P, Lee, W. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural modification of NADPH oxidase activator (Noxa 1) by oxidative stress: An experimental and computational study.
Int.J.Biol.Macromol., 163, 2020
|
|
5A2P
 
 | |
2D7J
 
 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D38
 
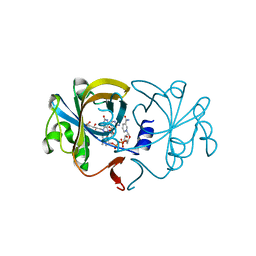 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
1MPV
 
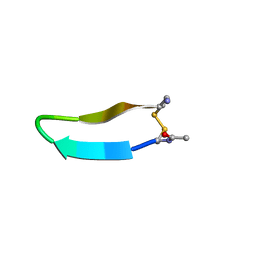 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | Descriptor: | BLyS Receptor 3 | | Authors: | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | Deposit date: | 2002-09-12 | | Release date: | 2002-10-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
1O07
 
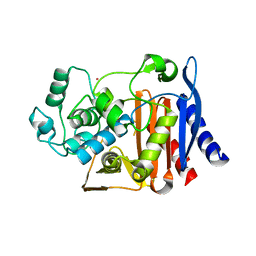 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | Descriptor: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | Authors: | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | Deposit date: | 2003-02-20 | | Release date: | 2003-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
1L3X
 
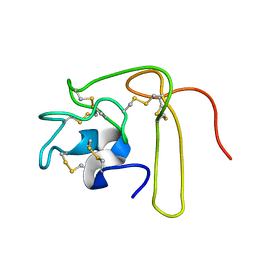 | | Solution Structure of Novel Disintegrin Salmosin | | Descriptor: | platelet aggregation inhibitor disintegrin | | Authors: | Shin, J, Lee, W. | | Deposit date: | 2002-03-01 | | Release date: | 2003-12-23 | | Last modified: | 2012-11-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel disintegrin, salmosin, from Agkistrondon halys venom
Biochemistry, 42, 2003
|
|
2N7Y
 
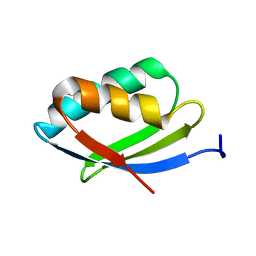 | | NMR structure of metal-binding domain 1 of ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Yu, C, Lee, W, Dmitriev, O. | | Deposit date: | 2015-09-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Metal Binding Domain 1 of the Copper Transporter ATP7B Reveals Mechanism of a Singular Wilson Disease Mutation.
Sci Rep, 8, 2018
|
|
1MJE
 
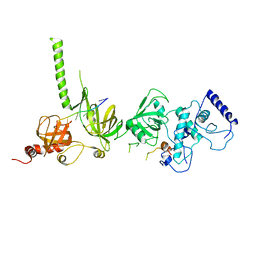 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
3W8L
 
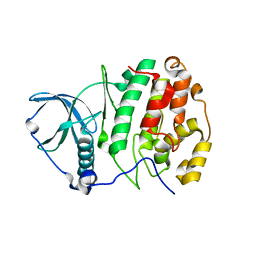 | | Crystal structure of human CK2 in complex with inositol hexakisphosphate | | Descriptor: | Casein kinase II subunit alpha, INOSITOL HEXAKISPHOSPHATE | | Authors: | Son, S.H, Lee, W.-K, Yu, Y.G, Lee, H.H. | | Deposit date: | 2013-03-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2EB1
 
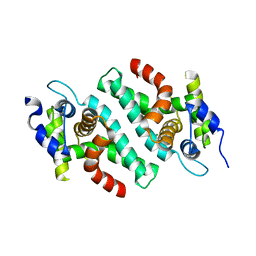 | | Crystal Structure of the C-Terminal RNase III Domain of Human Dicer | | Descriptor: | Endoribonuclease Dicer, MAGNESIUM ION | | Authors: | Takeshita, D, Zenno, S, Lee, W.C, Nagata, K, Saigo, K, Tanokura, M. | | Deposit date: | 2007-02-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homodimeric Structure and Double-stranded RNA Cleavage Activity of the C-terminal RNase III Domain of Human Dicer
J.Mol.Biol., 374, 2007
|
|
7V6U
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | The crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6S
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | The crystal structure fre
To be published
|
|
3GDC
 
 | |
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MONELLIN | | Authors: | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
1R02
 
 | |
2G1E
 
 | | Solution Structure of TA0895 | | Descriptor: | hypothetical protein Ta0895 | | Authors: | Yeo, I.Y, Hong, E, Jung, J, Lee, W, Yee, A, Arrowsmith, C.H. | | Deposit date: | 2006-02-14 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TA0895, a MoaD homologue from Thermoplasma acidophilum
Proteins, 65, 2006
|
|
