8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | Descriptor: | 3C-like proteinase nsp5, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8GLZ
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid. Crystal was initially co-crystallised with 4-methoxybenzoic acid and soaked with 4 mM hydrogen peroxide | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM2
 
 | |
8K05
 
 | |
8GLY
 
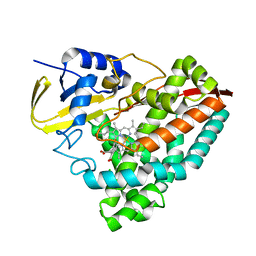 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
2VLG
 
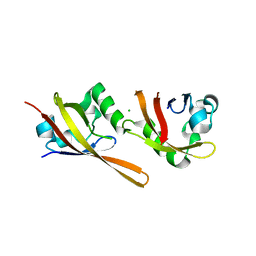 | | KinA PAS-A domain, homodimer | | Descriptor: | ACETATE ION, CHLORIDE ION, SPORULATION KINASE A | | Authors: | Lee, J, Tomchick, D.R, Brautigam, C.A, Machius, M, Kort, R, Hellingwerf, K.J, Gardner, K.H. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Changes at the Kina Pas-A Dimerization Interface Influence Histidine Kinase Function.
Biochemistry, 47, 2008
|
|
8GM1
 
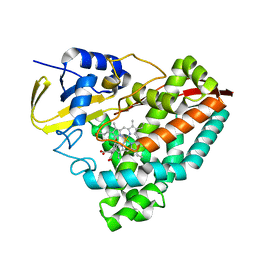 | | Crystal structure of T252E-CYP199A4 in complex with 4-methoxybenzoic acid soaked with 1 mM hydrogen peroxide | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
2V2W
 
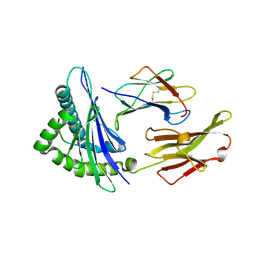 | | T CELL CROSS-REACTIVITY AND CONFORMATIONAL CHANGES DURING TCR ENGAGEMENT | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, Harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, Van Der Merwe, P.A, Jones, E.Y, Mcmichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
2V2X
 
 | | T cell cross-reactivity and conformational changes during TCR engagement. | | Descriptor: | BETA-2 MICROGLOBULIN, HIV P17, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Lee, J.K, Stewart-Jones, G, Dong, T, harlos, K, Di Gleria, K, Dorrell, L, Douek, D.C, van der Merwe, P.A, Jones, E.Y, McMichael, A.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T Cell Cross-Reactivity and Conformational Changes During Tcr Engagement.
J.Exp.Med., 200, 2004
|
|
1Z5P
 
 | | Crystal structure of MTA/AdoHcy nucleosidase with a ligand-free purine binding site | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5O
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5N
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
8WP9
 
 | | Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Lee, J, Ryu, B, Kim, T, Kim, K.K. | | Deposit date: | 2023-10-09 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of a 16.5-kDa small heat-shock protein from Methanocaldococcus jannaschii.
Int.J.Biol.Macromol., 258, 2024
|
|
7Y7O
 
 | |
6KHE
 
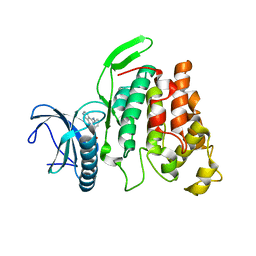 | | Crystal structure of CLK2 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
6KHF
 
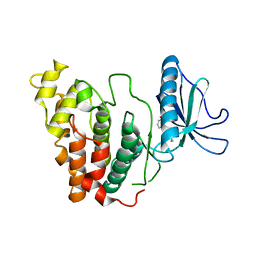 | | Crystal structure of CLK3 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK3 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
3QD6
 
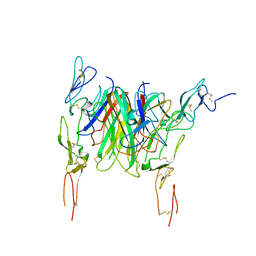 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
2Z80
 
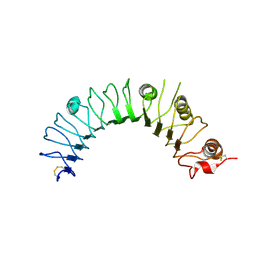 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 2, Variable lymphocyte receptor B | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
2Z81
 
 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 2, ... | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
