3ZTH
 
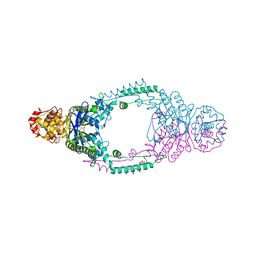 | | Crystal structure of Stu0660 of Streptococcus thermophilus | | Descriptor: | MAGNESIUM ION, STU0660 | | Authors: | Lee, K.-H, Robinson, H, Oh, B.-H. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, Structural, and Biochemical Characterization of a Group of Large Csn2 Proteins Involved in Crispr-Mediated Bacterial Immunity.
Proteins, 80, 2012
|
|
7LLQ
 
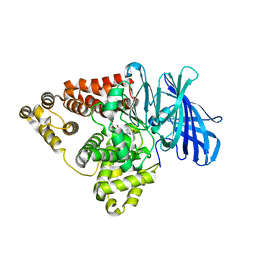 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
8YPJ
 
 | |
6L25
 
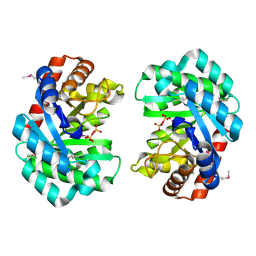 | | Deoxyribonuclease from Staphylococcus aureus | | Descriptor: | Deoxyribonuclease YcfH, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Lee, K.-Y, Kim, D.-G, Lee, B.-J. | | Deposit date: | 2019-10-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A structural study of TatD from Staphylococcus aureus elucidates a putative DNA-binding mode of a Mg2+-dependent nuclease.
Iucrj, 7, 2020
|
|
7C13
 
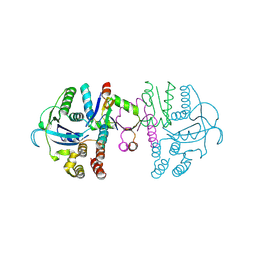 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin, Peptide methionine sulfoxide reductase MsrA | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C12
 
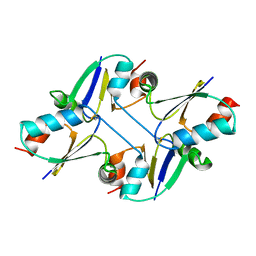 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C10
 
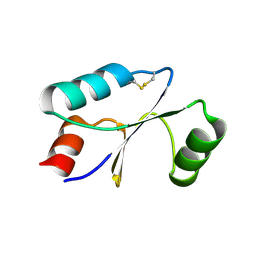 | | Dithiol cGrx1 | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
1CQ0
 
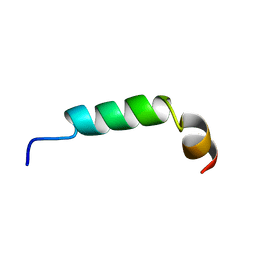 | | SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B'SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B ' | | Descriptor: | PROTEIN (NEW HYPOTHALAMIC NEUROPEPTIDE/OREXIN-B28) | | Authors: | Lee, K.-H, Bang, E.J, Chae, K.-J, Lee, D.W, Lee, W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-01-10 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B.
Eur.J.Biochem., 266, 1999
|
|
2WCV
 
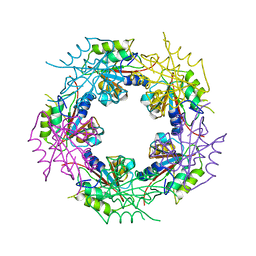 | | Crystal structure of bacterial FucU | | Descriptor: | L-FUCOSE MUTAROTASE, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
2WCU
 
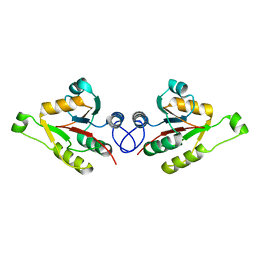 | | Crystal structure of mammalian FucU | | Descriptor: | PROTEIN FUCU HOMOLOG, alpha-L-fucopyranose | | Authors: | Lee, K.-H, Kim, M.-S, Suh, H.-Y, Ku, B, Song, Y.-L, Oh, B.-H. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures and Enzyme Mechanism of a Dual Fucose Mutarotase/Ribose Pyranase
J.Mol.Biol., 391, 2009
|
|
1P0L
 
 | | HP (2-20) Substitution GLN To TRP Modification In SDS-D25 MICELLES | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P5L
 
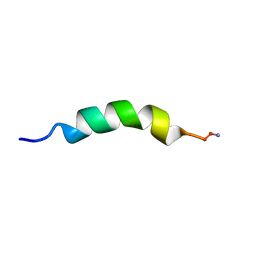 | | HP (2-20) Substitution PHE5 to SER modification in sds-d25 micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
8TWX
 
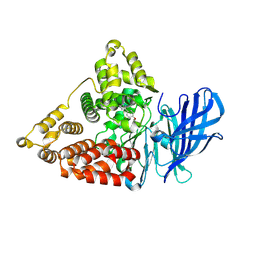 | |
1P0J
 
 | | HP (2-20) Substitution ASP To TRP Modification In SDS-D25 Micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P5K
 
 | | HP (2-20) Substitution SER to LEU11 modification in sds-d25 micelles | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-27 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P0G
 
 | | Structure of Antimicrobial Peptide, HP (2-20) and its Analogues Derived from Helicobacter pylori, as Determined by 1H NMR Spectroscopy | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
1P0O
 
 | | HP (2-20) substitution of Trp for Gln and Asp at position 17 and 19 MODIFICATION IN SDS-D25 MICELLES | | Descriptor: | 19-mer peptide from 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y.K, Harm, K.S, Kim, Y.M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Interactions between antimicrobial peptide, HP(2-20) derived from helicobacter pylori, and membrain studied by nmr spectroscopy
To be published
|
|
4AKF
 
 | |
1OT0
 
 | | Structure of Antimicrobial Peptide, HP (2-20) and its Analogues Derived from Helicobacter pylori, as Determined by 1H NMR Spectroscopy | | Descriptor: | 50S ribosomal protein L1 | | Authors: | Lee, K.H, Lee, D.G, Park, Y, Hahm, K.-S, Kim, Y. | | Deposit date: | 2003-03-21 | | Release date: | 2004-09-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of Antimicrobial Peptide, HP (2-20) and its Analogues Derived from Helicobacter pylori, as Determined by 1H NMR Spectroscopy
To be published
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4E
 
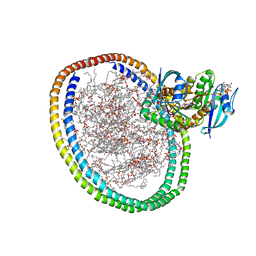 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7M5Y
 
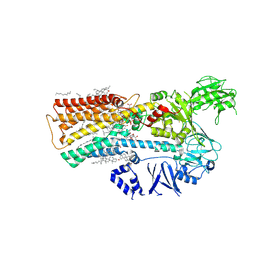 | | human ATP13A2 in the outward-facing E2 state bound to spermine and magnesium fluoride | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lee, K.P.K. | | Deposit date: | 2021-03-25 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanisms for gating and ion selectivity of the human polyamine transporter ATP13A2.
Mol.Cell, 81, 2021
|
|
7M5V
 
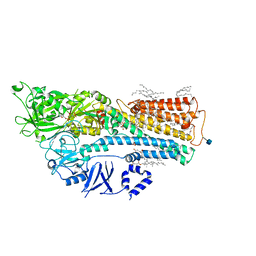 | | human ATP13A2 in the AMPPNP-bound occluded state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 1-DODECANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, K.P.K. | | Deposit date: | 2021-03-24 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms for gating and ion selectivity of the human polyamine transporter ATP13A2.
Mol.Cell, 81, 2021
|
|
7M5X
 
 | | Human ATP13A2 in the outward-facing E2 state bound to spermine and beryllium fluoride | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Lee, K.P.K. | | Deposit date: | 2021-03-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanisms for gating and ion selectivity of the human polyamine transporter ATP13A2.
Mol.Cell, 81, 2021
|
|
7E3W
 
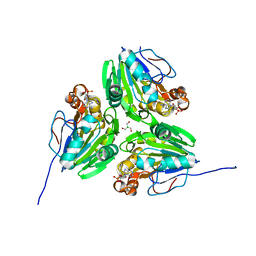 | | Metallo beta-lactamase fold protein (cAMP bound) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Lee, K.-Y, Kim, D.-G, Lee, B.-J. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional studies of SAV1707 from Staphylococcus aureus elucidate its distinct metal-dependent activity and a crucial residue for catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
