7XRI
 
 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7CPZ
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | BIOTIN, Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | Descriptor: | Mature Streptoavidin-C1 | | Authors: | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | Deposit date: | 2020-08-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
1WOG
 
 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOH
 
 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOI
 
 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F3D
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
7WG3
 
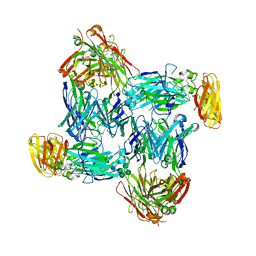 | | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody D9 for guiding humanization and affinity maturation for cancer therapy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D9 Fab, IL17RB protein, ... | | Authors: | Lee, W.H, Chen, X.R, Liu, I.J, Lee, J.H, Hu, C.M, Wu, H.C, Wang, S.K, Lee, W.H, Ma, C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody for guiding humanization and affinity maturation.
Cell Rep, 41, 2022
|
|
1BV2
 
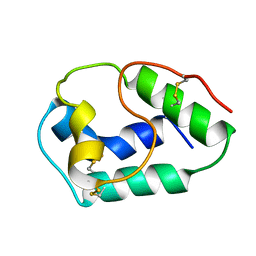 | | LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Poznanski, J, Sodano, P, Suh, S.W, Lee, J.Y, Ptak, M, Vovelle, F. | | Deposit date: | 1998-09-21 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Eur.J.Biochem., 259, 1999
|
|
5C6F
 
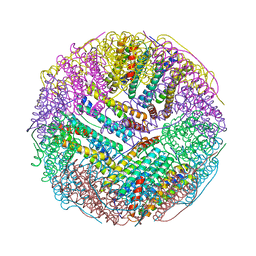 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
7XRH
 
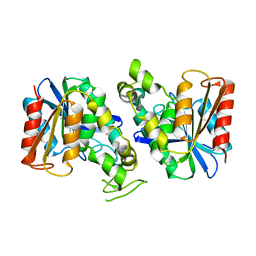 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
3A79
 
 | | Crystal structure of TLR2-TLR6-Pam2CSK4 complex | | Descriptor: | (2S)-propane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kang, J.Y, Jin, M.S, Lee, J.-O. | | Deposit date: | 2009-09-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer
Immunity, 31, 2009
|
|
7CRM
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-APO Structure | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside 2'-N-acetyltransferase | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CS0
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Paromomycin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
3RYO
 
 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
7YVT
 
 | |
3SXN
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5EPP
 
 | | Structural Insights into the Interaction of p97 N-terminus Domain and VBM Motif in Rhomboid Protease, RHBDL4 | | Descriptor: | Rhomboid-related protein 4, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Ly, T.T, Kang, J.Y, Lee, J.-G, An, J.Y, Youn, H.-S, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2015-11-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into the interaction of p97 N-terminus domain and VBM in rhomboid protease, RHBDL4.
Biochem.J., 473, 2016
|
|
4ZTT
 
 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
