2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
8Z7K
 
 | | Crystal structure of Hemolysin co-regulated protein 1 (Hcp1) VariantB from Burkholderia pseudomallei | | Descriptor: | Hcp family type VI secretion system effector, SULFATE ION | | Authors: | Lebedev, A.A, Charoenwattanasatien, R, Tandhavanant, S, Chantratita, N. | | Deposit date: | 2024-04-20 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Genetic variation of hemolysin co-regulated protein 1 affects the immunogenicity and pathogenicity of Burkholderia pseudomallei.
Plos Negl Trop Dis, 19, 2025
|
|
7ZSC
 
 | | Crystal structure of the heterodimeric human C-P4H-II with truncated alpha subunit (C-P4H-II delta281) | | Descriptor: | Prolyl 4-hydroxylase subunit alpha-2, Protein disulfide-isomerase, SULFATE ION | | Authors: | Lebedev, A, Koski, M.K, Wierenga, R.K, Murthy, A.V, Sulu, R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the collagen prolyl 4-hydroxylase (C-P4H) catalytic domain complexed with PDI: Toward a model of the C-P4H alpha 2 beta 2 tetramer.
J.Biol.Chem., 298, 2022
|
|
2C20
 
 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
2WNY
 
 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
3SS7
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
8FZV
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, expressed with SUMO tag | | Descriptor: | MAGNESIUM ION, Transcription factor ETV6,Anthrax toxin receptor 2, UNKNOWN ATOM OR ION | | Authors: | Pedroza Romo, M.J, Soleimani, S, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FZU
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, Expressed with SUMO tag | | Descriptor: | POTASSIUM ION, SULFATE ION, Transcription factor ETV6,Anthrax toxin receptor 2 | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5FYP
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, crystal form 2 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, PHOSPHOINOSITOL-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5FYR
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, myoinositol complex | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1AX4
 
 | | TRYPTOPHANASE FROM PROTEUS VULGARIS | | Descriptor: | POTASSIUM ION, TRYPTOPHANASE | | Authors: | Isupov, M.N, Antson, A.A, Dodson, E.J, Dodson, G.G, Dementieva, I.S, Zakomirdina, L.N, Wilson, K.S, Dauter, Z, Lebedev, A.A, Harutyunyan, E.H. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of tryptophanase.
J.Mol.Biol., 276, 1998
|
|
7NIT
 
 | | X-ray structure of a multidomain BbgIII from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Sanchez Rodriguez, F, Rigden, D.J, Tams, J.W, Wilting, R, Vester, J.K, Longhin, E, Krogh, K.B.R, Pache, R.A, Davies, G.J, Wilson, K.S. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7YLO
 
 | | Conversion of indole-3-acetic acid into indole-3-aldehyde in bacteria Metabolic network of tryptophan around the indole-3-aldehyde formation | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cheng, J, Luo, Y, Zhu, J, Tan, R, Wang, N, Lebedev, A.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular Insights into the One-Carbon Loss Oxidation of Indole-3-acetic Acid
Acs Catalysis, 14, 2024
|
|
6I7S
 
 | | Microsomal triglyceride transfer protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Biterova, E, Isupov, M.N, Keegan, R.M, Lebedev, A.A, Ruddock, L.W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of human microsomal triglyceride transfer protein.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H78
 
 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
5NPT
 
 | | Structure of the N-terminal domain of the yeast telomerase reverse transcriptase | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Rodina, E.V, Lebedev, A.A, Hakanpaa, J, Hackenberg, C, Petrova, O.A, Zvereva, M.I, Dontsova, O.A, Lamzin, V.S. | | Deposit date: | 2017-04-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase.
Nucleic Acids Res., 46, 2018
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
3RFZ
 
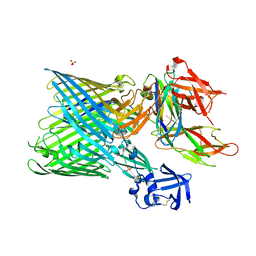 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
3O6Q
 
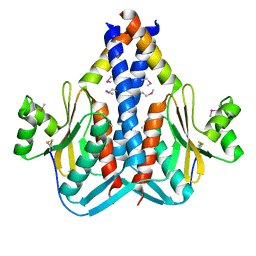 | | The Structure of SpoIISA and SpoIISB, a Toxin - Antitoxin System | | Descriptor: | Stage II sporulation protein SA, Stage II sporulation protein SB | | Authors: | Levdikov, V.M, Blagova, E.V, Lebedev, A.A, Wilkinson, A.J, Florek, P, Barak, I. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and interactions of SpoIISA and SpoIISB, a toxin-antitoxin system in Bacillus subtilis.
J.Biol.Chem., 161, 2010
|
|
2YHF
 
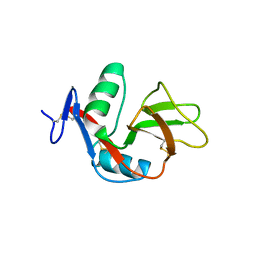 | | 1.9 Angstrom Crystal Structure of CLEC5A | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 5 MEMBER A | | Authors: | Watson, A.A, Lebedev, A.A, Murshudov, G.M, Vagin, A.A, Hall, B.A, O'Callaghan, C.A. | | Deposit date: | 2011-04-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Flexibility of the Macrophage Dengue Virus Receptor Clec5A: Implications for Ligand Binding and Signaling.
J.Biol.Chem., 286, 2011
|
|
5HT6
 
 | | Crystal structure of the SET domain of the human MLL5 methyltransferase | | Descriptor: | Histone-lysine N-methyltransferase 2E | | Authors: | le Maire, A, Mas-y-Mas, S, Dumas, C, Lebedev, A. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The Human Mixed Lineage Leukemia 5 (MLL5), a Sequentially and Structurally Divergent SET Domain-Containing Protein with No Intrinsic Catalytic Activity.
Plos One, 11, 2016
|
|
4CNU
 
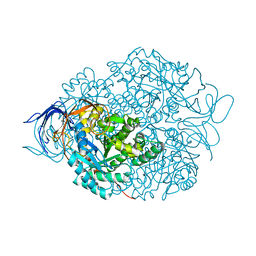 | |
3SS9
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
5DJS
 
 | | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M | | Descriptor: | Tetratricopeptide TPR_2 repeat protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
