8FXS
 
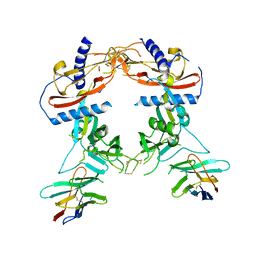 | | Crystal structure of human pro-TGF-beta2 in complex with Nb9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody clone 9, Transforming growth factor beta-2 proprotein | | Authors: | Le, V.Q, Springer, T.A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A specialized integrin-binding motif enables proTGF-beta 2 activation by integrin alpha V beta 6 but not alpha V beta 8.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FXV
 
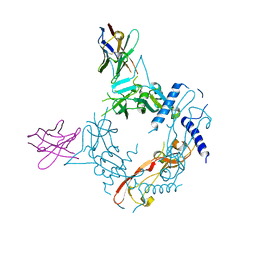 | | Crystal structure of human proTGF-beta2 in complex with Nb18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody clone 18, Transforming growth factor beta-2 proprotein | | Authors: | Le, V.Q, Springer, T.A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A specialized integrin-binding motif enables proTGF-beta 2 activation by integrin alpha V beta 6 but not alpha V beta 8.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2GDO
 
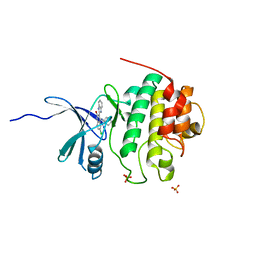 | | 4-(Aminoalkylamino)-3-Benzimidazole-Quinolinones As Potent CHK1 Inhibitors | | Descriptor: | 4-[(3S)-1-AZABICYCLO[2.2.2]OCT-3-YLAMINO]-3-(1H-BENZIMIDAZOL-2-YL)-6-CHLOROQUINOLIN-2(1H)-ONE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Le, V, Dove, J, Fang, E, Bussiere, D.E. | | Deposit date: | 2006-03-16 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7KWX
 
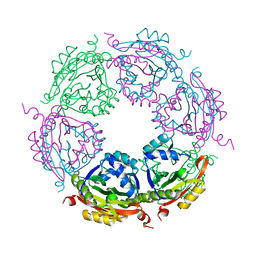 | | Spermidine N-acetyltransferase SpeG N152L mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KWQ
 
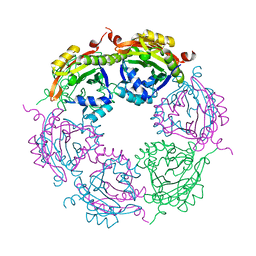 | | Spermidine N-acetyltransferase SpeG R149-K152 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KWJ
 
 | | Spermidine N-acetyltransferase SpeG K23-Q34 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KX3
 
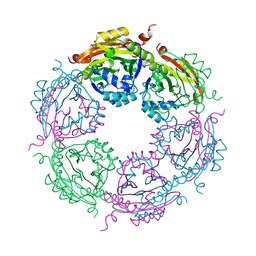 | | SpeG Spermidine N-acetyltransferase F149G mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KX2
 
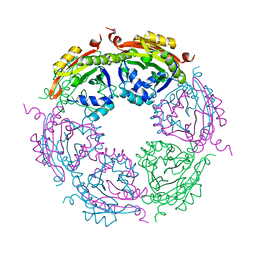 | | Spermidine N-acetyltransferase SpeG F149A mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
7KWH
 
 | | Spermidine N-acetyltransferase SpeG K23-Y30 chimera from Vibrio cholerae and hSSAT | | Descriptor: | PHOSPHATE ION, Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
4YCI
 
 | | non-latent pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, ... | | Authors: | Mi, L.Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6VFN
 
 | | Crystal structure of SpeG allosteric polyamine acetyltransferase from Bacillus thuringiensis in complex with spermine | | Descriptor: | SPERMINE, Spermidine N1-acetyltransferase | | Authors: | Tsimbalyuk, S, Shornikov, A, Le, V.T.B, Kuhn, M.L, Forwood, J.K. | | Deposit date: | 2020-01-05 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency.
J.Struct.Biol., 210, 2020
|
|
4YCG
 
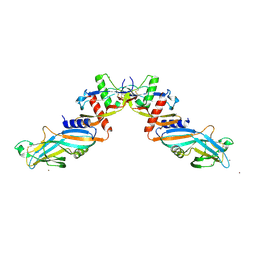 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6VFM
 
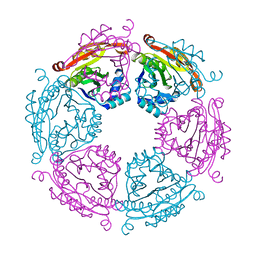 | | Crystal structure of SpeG allosteric polyamine acetyltransferase from Bacillus thuringiensis | | Descriptor: | Spermidine N1-acetyltransferase | | Authors: | Tsimbalyuk, S, Shornikov, A, Le, V.T.B, Kuhn, M.L, Forwood, J.K. | | Deposit date: | 2020-01-05 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | SpeG polyamine acetyltransferase enzyme from Bacillus thuringiensis forms a dodecameric structure and exhibits high catalytic efficiency.
J.Struct.Biol., 210, 2020
|
|
1B9F
 
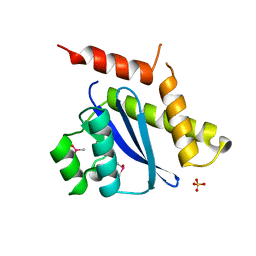 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
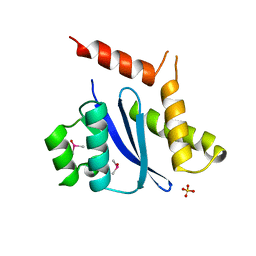 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
4N6Y
 
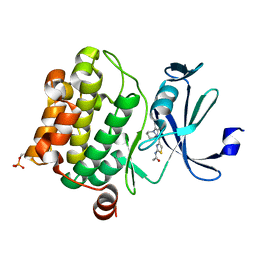 | | Pim1 Complexed with a phenylcarboxamide | | Descriptor: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4N6Z
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4N70
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | N-{4-[(3R,4R,5S)-3-amino-4-hydroxy-5-methylpiperidin-1-yl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
3K3B
 
 | | Co-crystal structure of the human kinesin Eg5 with a novel tetrahydro-beta-carboline | | Descriptor: | 3-[(1R)-2-acetyl-6-methyl-2,3,4,9-tetrahydro-1H-beta-carbolin-1-yl]phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Bussiere, D.E, Bellamacina, C, Le, V. | | Deposit date: | 2009-10-02 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The discovery of tetrahydro-beta-carbolines as inhibitors of the kinesin Eg5.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3T9I
 
 | | Pim1 complexed with a novel 3,6-disubstituted indole at 2.6 Ang Resolution | | Descriptor: | 2-methoxy-4-(3-phenyl-2H-pyrazolo[3,4-b]pyridin-6-yl)phenol, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Shu, W, Le, V, Nishiguchi, G, Bussiere, D. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-12 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel 3,5-disubstituted indole derivatives as potent inhibitors of Pim-1, Pim-2, and Pim-3 protein kinases.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
