1CX8
 
 | | CRYSTAL STRUCTURE OF THE ECTODOMAIN OF HUMAN TRANSFERRIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SAMARIUM (III) ION, TRANSFERRIN RECEPTOR PROTEIN | | Authors: | Lawrence, C.M, Ray, S, Babyonyshev, M, Galluser, R, Borhani, D, Harrison, S.C. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain of human transferrin receptor.
Science, 286, 1999
|
|
7STW
 
 | |
1SKV
 
 | | Crystal Structure of D-63 from Sulfolobus Spindle Virus 1 | | Descriptor: | Hypothetical 7.5 kDa protein | | Authors: | Kraft, P, Kummel, D, Oeckinghaus, A, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of d-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function
J.Virol., 78, 2004
|
|
6W11
 
 | |
2CO5
 
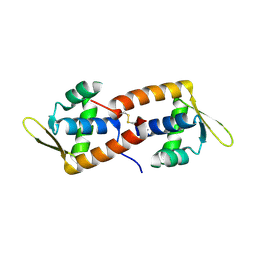 | | F93 FROM STIV, a winged-helix DNA-binding protein | | Descriptor: | VIRAL PROTEIN F93 | | Authors: | Larson, E.T, Reiter, D, Lawrence, C.M. | | Deposit date: | 2006-05-25 | | Release date: | 2006-05-31 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Winged-Helix Protein from Sulfolobus Turreted Icosahedral Virus Points Toward Stabilizing Disulfide Bonds in the Intracellular Proteins of a Hyperthermophilic Virus.
Virology, 368, 2007
|
|
2CLB
 
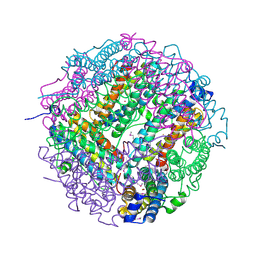 | | The structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like di-metal binding site within a Dps- like dodecameric assembly | | Descriptor: | DPS-LIKE PROTEIN, FE (III) ION, ZINC ION | | Authors: | Gauss, G.H, Benas, P, Wiedenheft, B, Young, M, Douglas, T, Lawrence, C.M. | | Deposit date: | 2006-04-26 | | Release date: | 2006-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Dps-Like Protein from Sulfolobus Solfataricus Reveals a Bacterioferritin-Like Dimetal Binding Site within a Dps-Like Dodecameric Assembly.
Biochemistry, 45, 2006
|
|
2SEB
 
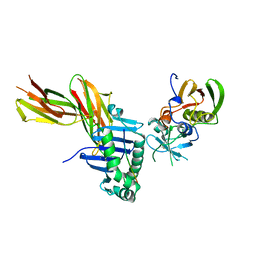 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH A PEPTIDE FROM HUMAN COLLAGEN II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Dessen, A, Lawrence, C.M, Cupo, S, Zaller, D.M, Wiley, D.C. | | Deposit date: | 1997-10-16 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II.
Immunity, 7, 1997
|
|
3UXU
 
 | |
8FO0
 
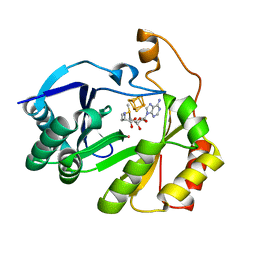 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FOL
 
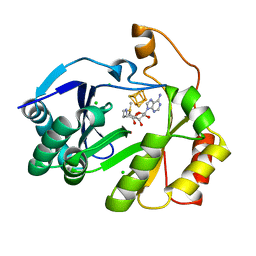 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FSI
 
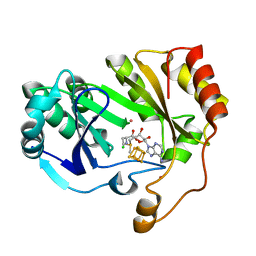 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
3PS0
 
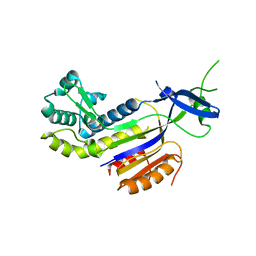 | | The structure of the CRISPR-associated protein, csa2, from Sulfolobus solfataricus | | Descriptor: | CRISPR-Associated protein, CSA2 | | Authors: | Lintner, N.G, Sdano, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE).
J.Biol.Chem., 286, 2011
|
|
3RKL
 
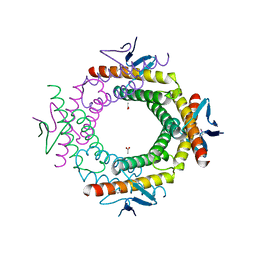 | |
3J31
 
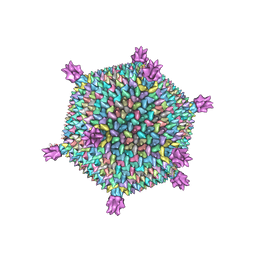 | | Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus | | Descriptor: | A223 penton base, A55 membrane protein, C381 turret protein, ... | | Authors: | Veesler, D, Ng, T.S, Sendamarai, A.K, Eilers, B.J, Lawrence, C.M, Lok, S.M, Young, M.J, Johnson, J.E, Fu, C.-Y. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6BO3
 
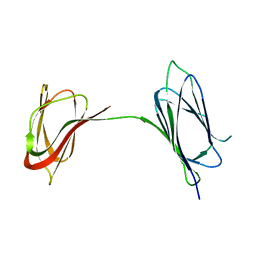 | | Structure Determination of A223, a turret protein in Sulfolobus turreted icosahedral virus, using an iterative hybrid approach | | Descriptor: | Uncharacterized protein | | Authors: | Sendamarai, A.K, Veesler, D, Fu, C.Y, Marceau, C, Larson, E.T, Johnson, J.E, Lawrence, C.M. | | Deposit date: | 2017-11-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A223, A STRUCTURAL PROTEIN FROM SULFOLOBUS TURRETED ICOSAHEDRAL VIRUS (STIV)
To Be Published
|
|
4IL7
 
 | |
4IND
 
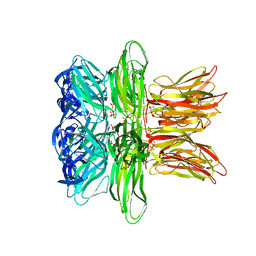 | | The Triple Jelly Roll Fold and Turret Assembly in an Archaeal Virus | | Descriptor: | C381 turret protein, DIHYDROGENPHOSPHATE ION | | Authors: | Eilers, B.J, Kraft, D, Burgess, M.C, Young, M.J, Lawrence, C.M. | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Triple Jelly Roll Fold and Turret Assembly in an Archaeal Virus
To be Published
|
|
4LID
 
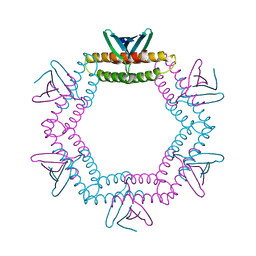 | | A100, A DNA binding scaffold from Sulfolobus spindle-shape virus 1 | | Descriptor: | A-100 | | Authors: | Eilers, B.J, Wagner, C, Thomas, M.M, Lawrence, C.M, Young, M.J. | | Deposit date: | 2013-07-02 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A100, A DNA binding scaffold from Sulfolobus spindle-shape virus 1
To be Published
|
|
1U6A
 
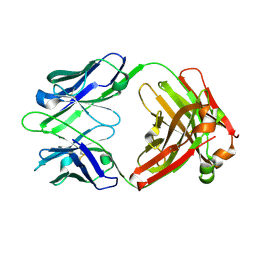 | | Crystal Structure of the Broadly Neutralizing Anti-HIV Fab F105 | | Descriptor: | F105 HEAVY CHAIN, F105 LIGHT CHAIN | | Authors: | Wilkinson, R.A, Piscitelli, C, Teintze, M, Lawrence, C.M. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120.
J.Virol., 79, 2005
|
|
2W8M
 
 | |
2WBT
 
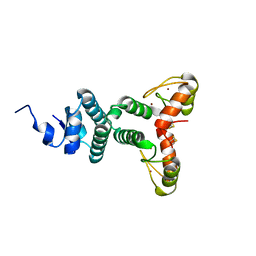 | | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA | | Descriptor: | B-129, ZINC ION | | Authors: | Eilers, B.J, Menon, S, Windham, A.B, Kraft, P, Dlakic, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of a Double C2H2 Zinc Finger Protein from a Hyperthermophilic Archaeal Virus in the Absence of DNA
To be Published
|
|
2VQ3
 
 | | Crystal Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferric Reductase of the Erythroid Transferrin Cycle | | Descriptor: | CITRIC ACID, METALLOREDUCTASE STEAP3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sendamarai, A.K, Ohgami, R.S, Fleming, M.D, Lawrence, C.M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferrireductase of the Erythroid Transferrin Cycle
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VZB
 
 | | A Dodecameric Thioferritin in the Bacterial Domain, Characterization of the Bacterioferritin-Related Protein from Bacteroides fragilis | | Descriptor: | BENZAMIDINE, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gauss, G.H, Young, M.J, Douglas, T, Lawrence, C.M. | | Deposit date: | 2008-07-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the Bacteroides Fragilis Bfr Gene Product Identifies a Bacterial Dps-Like Protein and Suggests Evolutionary Links in the Ferritin Superfamily.
J.Bacteriol., 194, 2012
|
|
1TBX
 
 | | Crystal structure of SSV1 F-93 | | Descriptor: | Hypothetical 11.0 kDa protein | | Authors: | Kraft, P, Oeckinghaus, A, Kummel, D, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-05-20 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein.
J.Virol., 78, 2004
|
|
2VNS
 
 | | Crystal Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferric Reductase of the Erythroid Transferrin Cycle | | Descriptor: | CITRIC ACID, METALLOREDUCTASE STEAP3 | | Authors: | Sendamarai, A.K, Ohgami, R.S, Fleming, M.D, Lawrence, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferrireductase of the Erythroid Transferrin Cycle
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
