6FMF
 
 | | Neuropilin-1 b1 domain in complex with EG01377; 2.8 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1, trifluoroacetic acid | | Authors: | Yelland, T, Djordjevic, S, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6FMC
 
 | | Neuropilin1-b1 domain in complex with EG01377, 0.9 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | Authors: | Yelland, T, Djordjevic, S, Fotinou, K, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
3ZIW
 
 | | Clostridium perfringens enterotoxin, D48A mutation and N-terminal 37 residues deleted | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
3ZIX
 
 | | Clostridium perfringens Enterotoxin with the N-terminal 37 residues deleted | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
6H6A
 
 | | Crystal structure of UNC119 in complex with LCK peptide | | Descriptor: | GLY-CYS-GLY-CYS-SER-SER, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Yelland, T, ElMaghloob, Y, McIlwraith, M, Stephen, L, Ismail, S. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-26 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK.
Dev.Cell, 47, 2018
|
|
2YBT
 
 | | Crystal structure of human acidic chitinase in complex with bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), ACIDIC MAMMALIAN CHITINASE, GLYCEROL | | Authors: | Sutherland, T.E, Andersen, O.A, Betou, M, Eggleston, I.M, Maizels, R.M, van Aalten, D, Allen, J.E. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor.
Chem. Biol., 18, 2011
|
|
7K7R
 
 | | EBNA1 peptide AA386-405 with Fab MS39p2w174 | | Descriptor: | CHLORIDE ION, EBNA1 peptide AA386-405, Fab HC MS39p2w174, ... | | Authors: | Lanz, T.V, Robinson, W.H, Jude, K.M. | | Deposit date: | 2020-09-23 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM.
Nature, 603, 2022
|
|
2YBU
 
 | | Crystal structure of human acidic chitinase in complex with bisdionin F | | Descriptor: | 3,7-DIMETHYL-1-[3-(3-METHYL-2,6-DIOXO-9H-PURIN-1-YL)PROPYL]PURINE-2,6-DIONE, ACIDIC MAMMALIAN CHITINASE, GLYCEROL | | Authors: | Sutherland, T.E, Andersen, O.A, Betou, M, Eggleston, I.M, Maizels, R.M, van Aalten, D, Allen, J.E. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor.
Chem. Biol., 18, 2011
|
|
1N0A
 
 | |
1SFK
 
 | | Core (C) protein from West Nile Virus, subtype Kunjin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | Authors: | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
1RQD
 
 | | deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form with the inhibitor GC7 bound in the active site | | Descriptor: | 1-GUANIDINIUM-7-AMINOHEPTANE, Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-12-04 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1RLZ
 
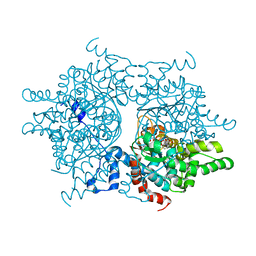 | | Deoxyhypusine synthase holoenzyme in its high ionic strength, low pH crystal form | | Descriptor: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-11-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1ROZ
 
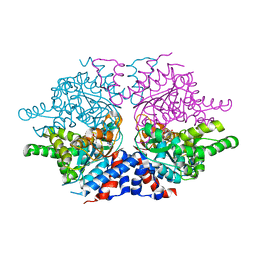 | | Deoxyhypusine synthase holoenzyme in its low ionic strength, high pH crystal form | | Descriptor: | Deoxyhypusine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Umland, T.C, Wolff, E.C, Park, M.-H, Davies, D.R. | | Deposit date: | 2003-12-02 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A New Crystal Structure of Deoxyhypusine Synthase Reveals the Configuration of the Active Enzyme and of an Enzyme-NAD-Inhibitor Ternary Complex
J.Biol.Chem., 279, 2004
|
|
1AF9
 
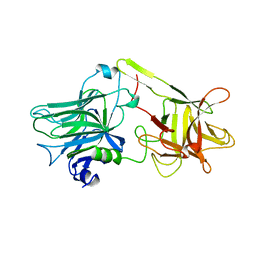 | | TETANUS NEUROTOXIN C FRAGMENT | | Descriptor: | TETANUS NEUROTOXIN | | Authors: | Umland, T.C, Wingert, L, Swaminathan, S, Furey, W.F, Schmidt, J.J, Sax, M. | | Deposit date: | 1997-03-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the receptor binding fragment HC of tetanus neurotoxin.
Nat.Struct.Biol., 4, 1997
|
|
1CCD
 
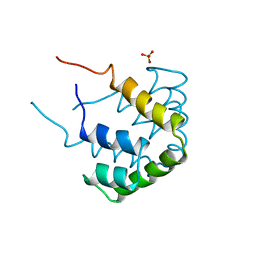 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
1CI4
 
 | |
1NGT
 
 | | The Role of Minor Groove Functional Groups in DNA Hydration | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(MTR)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Woods, K.K, Lan, T, McLaughlin, L.W, Williams, L.D. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Role of Minor Groove Functional Groups in DNA Hydration
Nucleic Acids Res., 31, 2003
|
|
1N09
 
 | | A minimal beta-hairpin peptide scaffold for beta-turn display | | Descriptor: | bhpW, disulfide cyclized beta-hairpin peptide | | Authors: | Russell, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stability of cyclic beta-hairpins: asymmetric contributions from side chains of a hydrogen-bonded cross-strand residue pair
J.Am.Chem.Soc., 125, 2003
|
|
1MX4
 
 | | Structure of p18INK4c (F82Q) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
6QF3
 
 | | X-Ray structure of Thermolysin soaked with sodium aspartate on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ASPARTIC ACID, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF4
 
 | | X-Ray structure of human Serine/Threonine Kinase 17B (STK17B) aka DRAK2 in complex with ADP obtained by on-chip soaking | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF5
 
 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
8EGS
 
 | |
4OV0
 
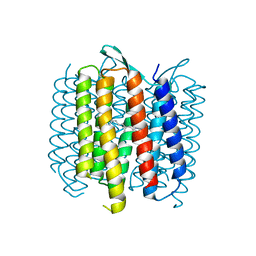 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | Deposit date: | 2014-02-19 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
