4L0J
 
 | | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems | | Descriptor: | DNA helicase I, MAGNESIUM ION, SULFATE ION | | Authors: | Redzej, A, Ilangovan, A, Lang, S, Gruber, C.J, Topf, M, Zangger, K, Zechner, E.L, Waksman, G. | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems.
Mol.Microbiol., 89, 2013
|
|
6I49
 
 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6FBU
 
 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | Descriptor: | ACETATE ION, DNA (5'-D(P*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3'), ... | | Authors: | Pomyalov, S, Lansky, S, Golan, G, Zharkov, D.O, Grollman, A.P, Shoham, G. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate
To Be Published
|
|
3ZWZ
 
 | | Crystal structure of Plasmodium falciparum AMA1 in complex with a 39aa PfRON2 peptide | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, AMA1, GLYCEROL, ... | | Authors: | Vulliez-Le Normand, B, Tonkin, M.L, Lamarque, M.H, Langer, S, Hoos, S, Roques, M, Saul, F.A, Faber, B.W, Bentley, G.A, Boulanger, M.J, Lebrun, M. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Insight Into the Malaria Parasite Moving Junction Complex
Plos Pathog., 8, 2012
|
|
7UG7
 
 | | 70S ribosome complex in an intermediate state of translocation bound to EF-G(GDP) stalled by Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Wieland, M, Holm, M, Koller, T.O, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1AJT
 
 | | FIVE-NUCLEOTIDE BULGE LOOP FROM TETRAHYMENA THERMOPHILA GROUP I INTRON, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*AP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*AP*AP*UP*AP*AP*GP*CP*UP*C)-3') | | Authors: | Luebke, K.J, Landry, S.M, Tinoco Junior, I. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a five-nucleotide RNA bulge loop from a group I intron.
Biochemistry, 36, 1997
|
|
2QYB
 
 | |
6VQP
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
6QCC
 
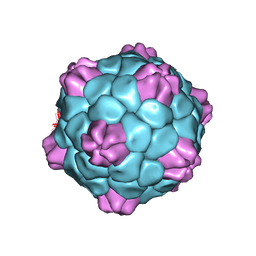 | | Cryo-EM Atomic Structure of Broad Bean Stain Virus (BBSV) | | Descriptor: | Large coat-protein subunit, Small coat-protein subunit | | Authors: | Lecorre, F, Lai Jee Him, J, Blanc, S, Zeddam, J.-L, Trapani, S, Bron, P. | | Deposit date: | 2018-12-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The cryo-electron microscopy structure of Broad Bean Stain Virus suggests a common capsid assembly mechanism among comoviruses.
Virology, 530, 2019
|
|
5DOX
 
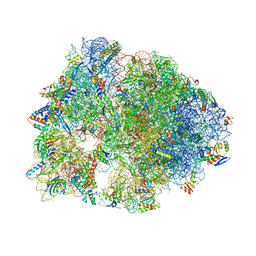 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
4V9D
 
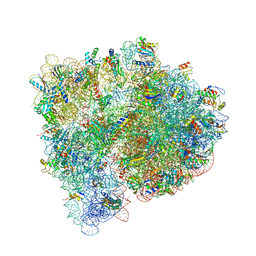 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
4UQZ
 
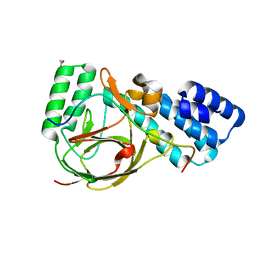 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | ACETATE ION, HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
RNA, 20, 2014
|
|
5C4Y
 
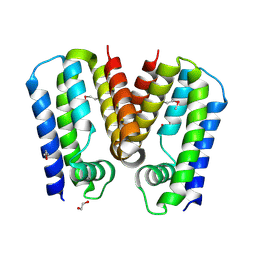 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
6EDG
 
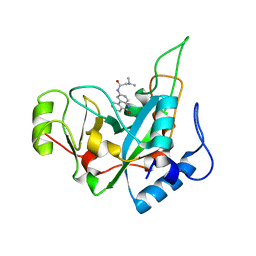 | | Pseudomonas exotoxin A domain III T18H477L | | Descriptor: | Exotoxin, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Moss, D.L, Park, H.W, Mettu, R.R, Landry, S.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Deimmunizing substitutions in Pseudomonasexotoxin domain III perturb antigen processing without eliminating T-cell epitopes.
J.Biol.Chem., 294, 2019
|
|
4R1H
 
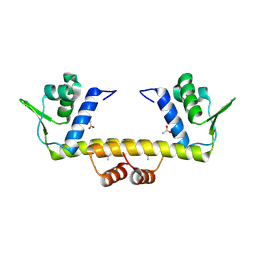 | |
6I47
 
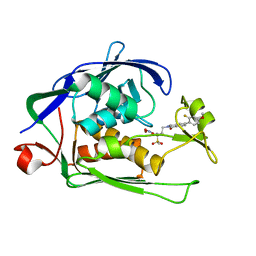 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
8QU8
 
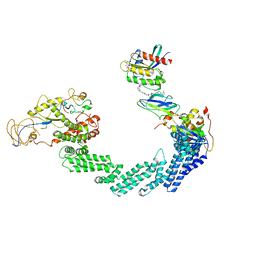 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
4PEV
 
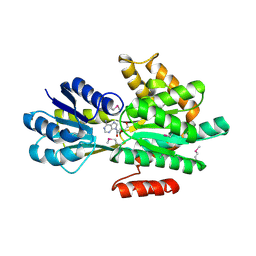 | | Crystal structure of ABC transporter system solute-binding proteins from Aeropyrum pernix K1 | | Descriptor: | ADENOSINE, GLYCEROL, Membrane lipoprotein family protein | | Authors: | Chang, C, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of ABC transporter system solute-binding proteins from Aeropyrum pernix K1
to be published
|
|
8FU7
 
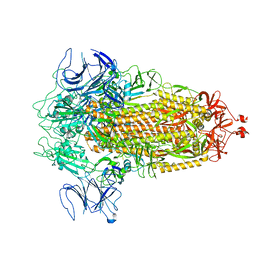 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
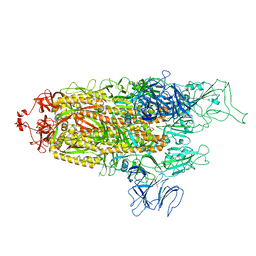 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
4RGP
 
 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | Descriptor: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
1B8W
 
 | | DEFENSIN-LIKE PEPTIDE 1 | | Descriptor: | PROTEIN (DEFENSIN-LIKE PEPTIDE 1) | | Authors: | Torres, A.M, Wang, X, Fletcher, J.I, Alewood, D, Alewood, P.F, Smith, R, Simpson, R.J, Nicholson, G.M, Sutherland, S.K, Gallagher, C.H, King, G.F, Kuchel, P.W. | | Deposit date: | 1999-02-02 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a defensin-like peptide from platypus venom.
Biochem.J., 341, 1999
|
|
2JHN
 
 | | 3-methyladenine dna-glycosylase from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-METHYLADENINE DNA-GLYCOSYLASE, GLYCEROL, ... | | Authors: | Leiros, I, Nabong, M.P, Grosvik, K, Ringvoll, J, Haugland, G.T, Uldal, L, Reite, K, Olsbu, I.K, Knaevelsrud, I, Moe, E, Andersen, O.A, Birkeland, N.K, Ruoff, P, Klungland, A, Bjelland, S. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Enzymatic Excision of N1-Methyladenine and N3-Methylcytosine from DNA
Embo J., 26, 2007
|
|
2R78
 
 | |
