4GZZ
 
 | | Crystal structures of bacterial RNA Polymerase paused elongation complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Weixlbaumer, A, Leon, K, Landick, R, Darst, S.A. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.2927 Å) | | Cite: | Structural basis of transcriptional pausing in bacteria.
Cell(Cambridge,Mass.), 152, 2013
|
|
4IAO
 
 | | Crystal structure of Sir2 C543S mutant in complex with SID domain of Sir4 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent histone deacetylase SIR2, Regulatory protein SIR4, ... | | Authors: | Hsu, H.C, Wang, C.L, Wang, M, Yang, N, Chen, Z, Sternglanz, R, Xu, R.M. | | Deposit date: | 2012-12-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding
Genes Dev., 27, 2013
|
|
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
3QWL
 
 | | Crystal structure of human TBC1 domain family member 7 | | Descriptor: | TBC1 domain family member 7, UNKNOWN ATOM OR ION | | Authors: | Guan, X, Tempel, W, Tong, Y, Wernimont, A.K, Landry, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human TBC1 domain family member 7
to be published
|
|
4BUR
 
 | | Crystal structure of the reduced human Apoptosis inducing factor complexed with NAD | | Descriptor: | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
4IUW
 
 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
1O7D
 
 | | The structure of the bovine lysosomal a-mannosidase suggests a novel mechanism for low pH activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Heikinheimo, P, Helland, R, Leiros, H.S, Leiros, I, Karlsen, S, Evjen, G, Ravelli, R, Schoehn, G, Ruigrok, R, Tollersrud, O.-K, Mcsweeney, S, Hough, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of Bovine Lysosomal Alpha-Mannosidase Suggests a Novel Mechanism for Low-Ph Activation
J.Mol.Biol., 327, 2003
|
|
1EJM
 
 | | CRYSTAL STRUCTURE OF THE BPTI ALA16LEU MUTANT IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | BETA-TRYPSIN, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Otlewski, J, Smalas, A, Helland, R, Grzesiak, A, Krowarsch, D. | | Deposit date: | 2000-03-03 | | Release date: | 2001-03-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substitutions at the P(1) position in BPTI strongly affect the association energy with serine proteinases.
J.Mol.Biol., 301, 2000
|
|
4N5H
 
 | | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Esterase/lipase, ... | | Authors: | Bennett, M.D, Holland, R, Loo, T.S, Smith, C.A, Norris, G.E, Delabre, M.-L, Anderson, B.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001)
TO BE PUBLISHED
|
|
1IB1
 
 | | CRYSTAL STRUCTURE OF THE 14-3-3 ZETA:SEROTONIN N-ACETYLTRANSFERASE COMPLEX | | Descriptor: | 14-3-3 ZETA ISOFORM, COA-S-ACETYL TRYPTAMINE, SEROTONIN N-ACETYLTRANSFERASE | | Authors: | Obsil, T, Ghirlando, R, Klein, D.C, Ganguly, S, Dyda, F. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation.
Cell(Cambridge,Mass.), 105, 2001
|
|
1P3Q
 
 | | Mechanism of Ubiquitin Recognition by the CUE Domain of VPS9 | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E.A, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2003-04-18 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p.
Cell(Cambridge,Mass.), 113, 2003
|
|
1PZ9
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1Q0Q
 
 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1PZ7
 
 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1QSM
 
 | | Histone Acetyltransferase HPA2 from Saccharomyces Cerevisiae in Complex with Acetyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, HPA2 HISTONE ACETYLTRANSFERASE | | Authors: | Angus-Hill, M.L, Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1999-06-22 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily.
J.Mol.Biol., 294, 1999
|
|
1Q0H
 
 | | Crystal structure of selenomethionine-labelled DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, CITRIC ACID, ... | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1Q0L
 
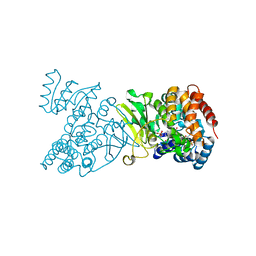 | | Crystal structure of DXR in complex with fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
1Q56
 
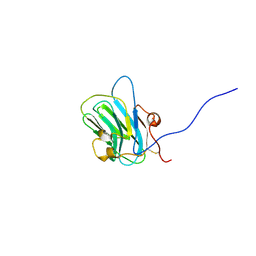 | | NMR structure of the B0 isoform of the agrin G3 domain in its Ca2+ bound state | | Descriptor: | Agrin | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-08-06 | | Release date: | 2004-04-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding
Structure, 12, 2004
|
|
1PZ8
 
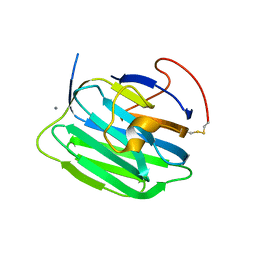 | | Modulation of agrin function by alternative splicing and Ca2+ binding | | Descriptor: | Agrin, CALCIUM ION | | Authors: | Stetefeld, J, Alexandrescu, A.T, Maciejewski, M.W, Jenny, M, Rathgeb-Szabo, K, Schulthess, T, Landwehr, R, Frank, S, Ruegg, M.A, Kammerer, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Modulation of agrin function by alternative splicing and Ca2+ binding.
STRUCTURE, 12, 2004
|
|
1RQQ
 
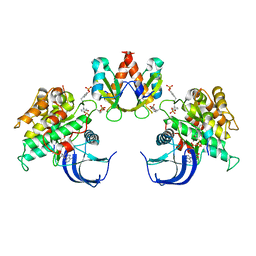 | | Crystal Structure of the Insulin Receptor Kinase in Complex with the SH2 Domain of APS | | Descriptor: | BISUBSTRATE INHIBITOR, Insulin receptor, MANGANESE (II) ION, ... | | Authors: | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | Deposit date: | 2003-12-06 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
1RPY
 
 | | CRYSTAL STRUCTURE OF THE DIMERIC SH2 DOMAIN OF APS | | Descriptor: | SULFATE ION, adaptor protein APS | | Authors: | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | Deposit date: | 2003-12-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
